UMass Amherst Chemists Create a Three-Color Tool to Watch RNA in Living Cells in Real Time
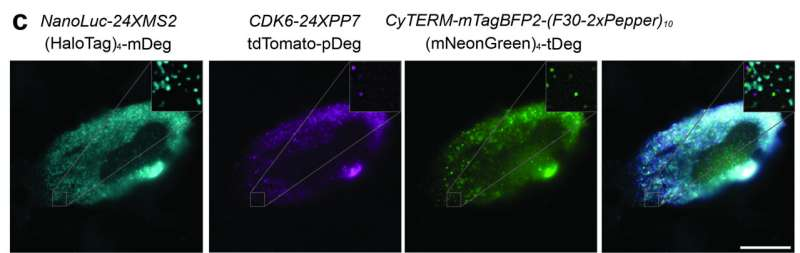
Chemists at the University of Massachusetts Amherst have developed a powerful new method that allows scientists to observe RNA molecules inside living mammalian cells with unprecedented clarity. The technique, which enables three-color imaging of mRNA in real time, represents a major step forward in understanding how RNA behaves, moves, and performs its many critical functions inside cells.
RNA is central to life. It carries instructions from DNA to make proteins, regulates gene activity, helps organize cellular structures, and plays key roles in health and disease. Despite its importance, RNA has remained notoriously difficult to study inside living cells, largely because of its tiny size and fast-moving nature. This new tool directly addresses that challenge.
The research was led by Daisy Pham, a graduate student in chemistry at UMass Amherst, and Jiahui (Chris) Wu, an assistant professor of chemistry. Their study was published in the journal Nature Methods, a leading outlet for cutting-edge experimental techniques in biology.
Why Studying RNA Inside Living Cells Is So Difficult
RNA molecules are small, dynamic, and constantly interacting with other cellular components. Traditional methods often require cells to be fixed or broken apart, which means researchers lose the ability to see how RNA behaves in its natural environment.
For years, scientists have relied on fluorescent tagging techniques to visualize RNA. One of the most widely used approaches is the RNA hairpin method, where fluorescent proteins are fused to RNA-binding elements that attach to specific RNA strands. While effective, this method has important limitations.
The biggest problem is background fluorescence. In many existing systems, fluorescent proteins glow all the time, whether or not they are bound to RNA. This constant glow can create visual noise that makes it harder to track individual RNA molecules accurately. Additionally, many tagging systems require relatively large RNA modifications, which can interfere with how RNA naturally functions.
A Smarter Way to Make RNA Visible
The UMass Amherst team built on existing RNA imaging strategies but introduced several crucial innovations.
Instead of using fluorescent proteins that are always glowing, the researchers engineered RNA-regulated destabilization domains. These are protein tags that are unstable and rapidly degraded inside the cell unless they bind to their specific RNA target. Only when the protein “plugs into” the correct RNA strand does it stabilize and emit light.
This means fluorescence appears only when and where the RNA is present, dramatically reducing background noise and improving image clarity.
Even more impressively, the team developed three distinct destabilization domains, each paired with a different fluorescent protein. These glow in green, red, and far-red, allowing researchers to track three different RNA populations at the same time inside a single living cell.
This three-color capability is what truly sets the method apart.
What Makes the Three-Color System Special
Each of the engineered protein-RNA pairs is orthogonal, meaning they operate independently and do not interfere with one another. One RNA type will only activate its matching fluorescent protein, and not the others. This precision allows researchers to assign different colors to different RNA molecules based on their function or identity.
With this system, scientists can now observe:
- Where different RNAs are located within the cell
- How they move over time
- How multiple RNA species interact or behave simultaneously
Importantly, the RNA tags used in this method are shorter than those required in many older techniques, which helps ensure that RNA molecules continue to function normally. This focus on minimal RNA perturbation is a major strength of the approach.
Why This Matters for Health and Disease Research
RNA plays a role in many diseases, including cancer, neurodegenerative disorders, and genetic conditions caused by errors in gene regulation. When RNA molecules malfunction, the consequences can be severe.
By allowing researchers to watch RNA in action inside living cells, this tool opens the door to deeper insights into how diseases develop at the molecular level. Scientists can now study how RNA localization, timing, and interactions change under different conditions, including stress, mutation, or drug treatment.
The ability to compare multiple RNA species at once also makes it easier to explore complex regulatory networks, rather than studying one molecule in isolation.
Making the Tool Available to the Scientific Community
One of the most important aspects of this work is that the researchers have made their method publicly available. Other labs can adopt and adapt the system for their own studies, accelerating progress across many areas of biology and medicine.
The team sees this tool as an addition to the growing toolkit for live-cell imaging, complementing other approaches rather than replacing them. In combination with advances in microscopy and computational analysis, this method could help answer long-standing questions about how cells function at the molecular level.
A Closer Look at RNA and Why It Deserves Attention
RNA is often described as DNA’s messenger, but that description barely scratches the surface. Beyond messenger RNA (mRNA), cells contain many other RNA types that regulate gene expression, modify proteins, and maintain cellular structure.
Over the past decade, scientists have discovered that RNA molecules are involved in:
- Turning genes on and off
- Responding rapidly to environmental changes
- Organizing regions inside the cell without membranes
- Fine-tuning protein production
Because RNA activity is so dynamic, real-time observation inside living cells is essential for understanding these processes. Tools like the one developed at UMass Amherst help bridge the gap between molecular biology and live-cell behavior.
How This Fits Into the Bigger Picture of RNA Imaging
RNA imaging has been a rapidly evolving field. Some techniques rely on fluorescent dyes that bind RNA, while others use RNA aptamers or CRISPR-based systems. Each approach has trade-offs involving brightness, specificity, background noise, and ease of use.
What makes this new three-color system stand out is its balance of clarity, flexibility, and minimal disruption to natural RNA behavior. By combining RNA-regulated protein stability with multicolor imaging, the method delivers clean signals while preserving biological realism.
As researchers continue to refine live-cell imaging technologies, tools like this one are likely to play a central role in future discoveries.
Looking Ahead
The development of this three-color RNA imaging method represents more than just a technical achievement. It reflects a broader shift toward studying biology as it happens, rather than relying solely on static snapshots.
By making it possible to watch different RNAs perform their roles inside living cells, the UMass Amherst team has given scientists a new way to explore one of life’s most essential molecules. As the method is adopted and expanded, it could lead to new insights into cellular organization, gene regulation, and the molecular roots of disease.
Research Paper:
https://www.nature.com/articles/s41592-025-02905-x