University of Tennessee Researchers Develop a Powerful New Statistical Method for Single-Molecule Fluorescence Analysis
Researchers at the University of Tennessee, Knoxville have introduced a new statistical method that significantly improves how scientists analyze single-molecule fluorescence experiments, a critical technique used to study proteins and molecular processes inside living cells. The work, published in the Biophysical Journal in 2025, brings together expertise from mathematics and biochemistry to tackle long-standing challenges in fluorescence data analysis.
Single-molecule fluorescence experiments allow scientists to observe biological processes at an incredibly fine scale, sometimes following the behavior of individual protein complexes in real time. While these experiments generate valuable data, analyzing that data accurately has always been difficult. Noise, photobleaching effects, and reliance on simplifying assumptions often limit how much information researchers can extract. This new method aims to change that.
A Truly Interdisciplinary Collaboration
The research was led by Associate Professor Ioannis Sgouralis and Ph.D. student Chiara Mattamira from the Department of Mathematics, alongside Professor Francisco Barrera, Associate Professor Rajan Lamichhane, graduate student Alyssa Ward, and postdoctoral associate Sriram Tiruvadi Krishnan from the Department of Biochemistry & Cellular and Molecular Biology (BCMB).
What makes this work stand out is the tight integration of theory and experiment. The mathematical framework was developed hand-in-hand with real experimental data generated in the BCMB laboratories. Instead of testing theory on artificial datasets alone, the team ensured that their models remained grounded in real biological measurements.
This collaboration helped avoid unrealistic assumptions and led to statistical tools that are both mathematically rigorous and experimentally relevant.
Why Traditional Analysis Methods Fall Short
In standard single-molecule fluorescence analysis, researchers often rely on assumptions about molecular behavior, such as fixed kinetic models or expected step patterns. In many cases, scientists must manually inspect data to identify photobleaching steps, a process that can be time-consuming, subjective, and prone to error.
Another common issue is signal averaging. Many fluorescence techniques average signals from millions of molecules, which smooths out noise but also hides important details about how individual molecules behave. This approach can mask rare events or complex molecular dynamics that are essential for understanding biological function.
These limitations create a strong need for automated, assumption-free, and statistically sound methods that can extract meaningful insights from noisy experimental data.
How the New Statistical Method Works
The newly developed approach uses advanced Bayesian statistical techniques to analyze single-molecule fluorescence time series data. Instead of assuming a predefined molecular model, the method allows the data itself to guide the analysis.
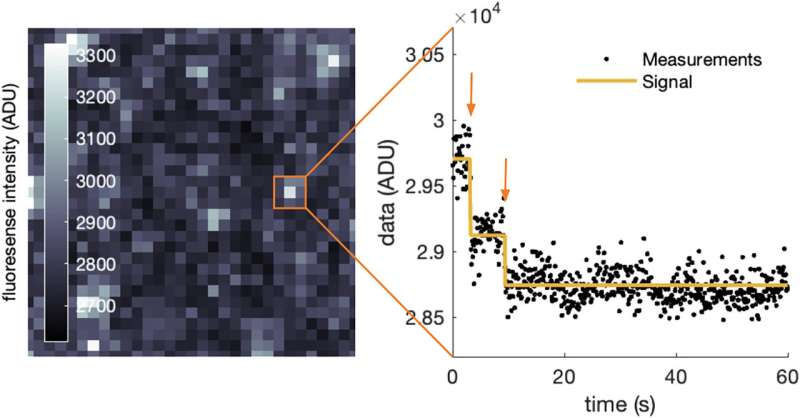
At its core, the method focuses on photobleaching step analysis, a common task in fluorescence experiments where researchers count discrete drops in fluorescence intensity as individual fluorophores bleach over time. Accurately identifying these steps is essential for determining molecular stoichiometry, such as how many subunits make up a protein complex.
The team designed efficient algorithms that can automatically infer:
- The number of photobleaching steps
- The timing of each step
- The uncertainty associated with each estimate
Importantly, the method performs well even under noisy conditions, where traditional approaches often struggle.
Tested on Real and Simulated Data
To validate their approach, the researchers applied the method to both simulated datasets and real experimental fluorescence data. Simulated data allowed them to test performance under controlled conditions, while real data demonstrated that the method works in practical laboratory settings.
Across both types of data, the new method consistently delivered accurate and reliable results, showing strong resistance to noise and experimental variability. This versatility makes it suitable for a wide range of fluorescence experiments and biological systems.
Why This Matters for Biological Research
Single-molecule fluorescence is widely used in biophysics, biochemistry, molecular biology, and structural biology. Improvements in data analysis directly translate into better understanding of:
- Protein complex assembly
- Molecular interactions
- Cellular dynamics in living systems
By reducing reliance on assumptions and manual interpretation, the new method helps researchers extract more information from the same experiments, increasing efficiency without disturbing sensitive biological samples.
Instead of averaging signals and losing detail, scientists can now focus on individual molecular behaviors, gaining clearer insight into complex biological processes.
Broader Impact and Future Applications
The research team believes this method has strong potential for global adoption among scientists who rely on single-molecule fluorescence techniques. As experimental technologies continue to improve, datasets are becoming larger and more complex, increasing the demand for robust analytical tools.
This method offers a way to process large volumes of data more efficiently while maintaining statistical reliability. It also opens the door to studying systems that were previously too noisy or complex to analyze accurately.
Training the Next Generation of Scientists
Beyond its technical impact, the project also highlights the value of interdisciplinary training. For graduate students and postdoctoral researchers involved, working across mathematics and experimental biology provided hands-on experience in collaboration, communication, and problem-solving.
Developing a shared language between disciplines helped transform abstract mathematical ideas into practical tools that experimental scientists can actually use. These skills are increasingly important as modern research continues to blur traditional academic boundaries.
A Closer Look at Single-Molecule Fluorescence
Single-molecule fluorescence experiments use light to track individual biomolecules that would otherwise be invisible. Unlike bulk techniques, which observe averaged behavior, single-molecule methods reveal heterogeneity, showing how different molecules behave differently under the same conditions.
However, this power comes with challenges. Signals are weak, noise is high, and photobleaching is unavoidable. Advances like this new statistical method are essential for fully unlocking the potential of these experiments.
What Comes Next
As more laboratories adopt model-free statistical approaches, the quality and reproducibility of fluorescence research are likely to improve. This work sets a strong example of how mathematical innovation can directly enhance experimental science.
By combining theory, computation, and biology, the University of Tennessee team has delivered a tool that not only solves a technical problem but also pushes the field forward.
Research paper:
https://doi.org/10.1016/j.bpj.2025.08.014