Scientists Discover How Gene Transcription Machinery Actively Shapes Aging and Cellular Senescence
Researchers at Northwestern Medicine have uncovered a detailed molecular link between gene expression and aging, showing that the way cells transcribe DNA into RNA plays a central role in whether cells continue dividing, pause their growth, or enter a state known as cellular senescence. The findings come from a new study published in the journal Molecular Cell in 2025 and add an important new layer to our understanding of how aging unfolds at the cellular level.
At the heart of the study is the process of transcription, where DNA is copied into messenger RNA (mRNA) so that proteins can be produced. While transcription has long been known to be essential for life, its precise role in aging—and particularly how transcription is regulated during aging—has remained relatively underexplored. This new research changes that.
The study was led by Ali Shilatifard, chair and Robert Francis Furchgott Professor of Biochemistry and Molecular Genetics at Northwestern University Feinberg School of Medicine, along with Saeid Parast, a postdoctoral research fellow and the study’s first author.
Understanding Cellular Senescence and Why It Matters
Cellular senescence is a state in which cells stop dividing but remain metabolically active. These cells do not die, but they also do not contribute to tissue renewal. Instead, they often release inflammatory signals and other factors that can damage surrounding tissues. Senescence is strongly associated with aging, tissue degeneration, and age-related diseases.
One of the long-standing questions in aging biology is whether senescence is a permanent endpoint or whether it can be reversed or modulated. This study provides strong evidence that senescence is not necessarily a one-way street.
The Role of Transcription Elongation Factors
The researchers focused on a group of proteins called transcription elongation factors. These proteins help RNA polymerase II, the enzyme responsible for synthesizing mRNA, move smoothly along DNA during transcription. Without proper elongation, gene expression becomes disrupted.
Using advanced genetic tools and high-resolution RNA sequencing, the team examined how specific elongation factors influence gene expression programs associated with senescence.
Two factors emerged as especially important: NELF (Negative Elongation Factor) and SPT6.
NELF and SPT6 as Key Regulators of Aging-Linked Gene Expression
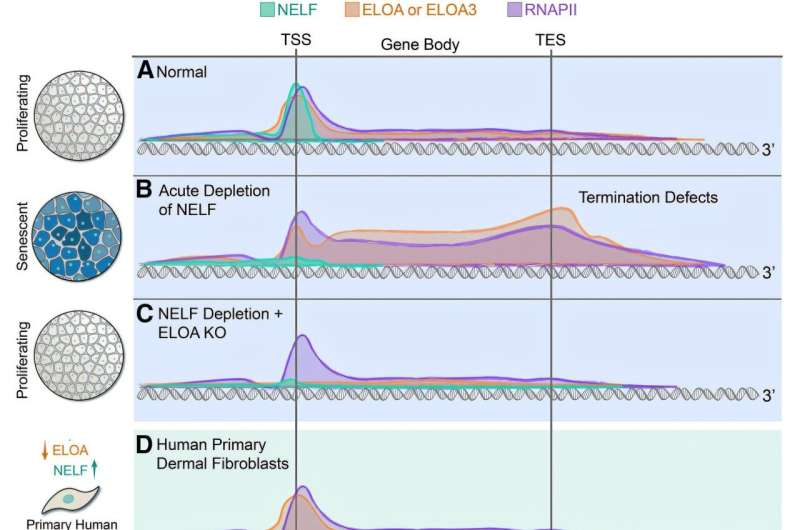
Previous work from Shilatifard’s laboratory had shown that deleting NELF caused cells to stop growing without killing them, pushing them into a senescent-like state. In the new study, researchers expanded on this observation to understand which genes and pathways were responsible for this growth arrest and whether cells could recover.
The researchers found that NELF and SPT6 regulate different aspects of transcription, particularly influencing which RNA isoforms are produced. RNA isoforms are alternative versions of mRNA generated from the same gene, and they can dramatically affect how a gene functions.
When NELF or SPT6 were depleted for extended periods, cells activated senescence-associated gene programs and temporarily stopped dividing. Importantly, this growth arrest was reversible once normal transcriptional control was restored. This finding strongly suggests that senescence can be actively regulated by transcription machinery, rather than being a permanently fixed state.
A Closer Look at RNA Processing and Isoforms
One of the study’s major insights is that aging-related changes are not only about which genes are turned on or off, but also about how genes are processed into RNA. The researchers observed widespread defects in RNA processing, leading to altered isoform usage in senescent cells.
These changes affect genes involved in stress responses, cell cycle regulation, and cellular maintenance—key systems that decline with age. By influencing isoform production, transcription elongation factors can subtly reshape cellular behavior without changing the underlying DNA sequence.
The Discovery of ELOA’s Role in Aging Cells
Beyond NELF and SPT6, the study identified another critical player: Elongin A (ELOA). Genetic screening revealed that ELOA plays a major role in transcription termination, the step where RNA polymerase stops copying DNA at the end of a gene.
ELOA was found to preferentially regulate short, stress-responsive genes, which are often activated rapidly when cells encounter environmental or internal stress. This selective regulation may reflect an evolutionary adaptation, allowing cells to respond quickly to threats.
When ELOA was removed, aging human fibroblasts—cells commonly used to study aging—showed a growth advantage, meaning they divided more readily. This suggests that ELOA normally acts as a molecular brake on cell proliferation during aging, reinforcing senescence-related growth arrest.
The Significance of ELOA3 and Genetic Variation
The researchers also highlighted ELOA3, a primate-specific homolog of ELOA that the group had characterized previously. ELOA3 contains a repeat cluster with variable repeat numbers across the human population.
This variability raises the possibility that natural genetic differences in ELOA3 could influence individual susceptibility to age-related diseases. While this connection remains speculative, it opens new avenues for research into how genetic variation shapes aging outcomes in humans.
Why This Study Matters for Aging Research
Taken together, the findings reveal a previously unrecognized link between transcription elongation control and aging biology. Rather than being a passive background process, transcription regulation actively determines whether cells enter, maintain, or exit senescence.
This insight has major implications. If aging-related senescence can be modulated by targeting transcription elongation factors, it may be possible to develop therapies that promote healthier aging. Such approaches could potentially reduce the burden of age-associated diseases without forcing cells into uncontrolled growth.
Broader Context: Transcription and Aging
This study fits into a growing body of research showing that aging is deeply connected to gene expression fidelity. As organisms age, transcription becomes noisier, RNA processing less precise, and stress-response systems less coordinated.
By pinpointing specific molecular regulators like NELF, SPT6, and ELOA, the Northwestern team has provided concrete targets for future investigation. These findings also reinforce the idea that aging is not driven by a single pathway, but by interconnected molecular systems that can, at least in part, be fine-tuned.
What Comes Next
The researchers emphasize that much work remains to be done. Future studies will need to explore how these transcription factors interact with other aging-related pathways, how they function in different cell types, and whether they can be safely targeted in living organisms.
Still, the message from this study is clear: transcriptional elongation control is central to aging, and understanding it could reshape how scientists think about senescence, longevity, and age-related disease.
Research Paper Reference:
https://doi.org/10.1016/j.molcel.2025.11.021