Scientists Build Virtual Tissue Tools That Reveal How Cells Communicate in Disease
Scientists at Duke-NUS Medical School have introduced two powerful new computational tools that could significantly change how researchers study the way cells communicate inside the body. These tools, named sCCIgen and QuadST, are designed to uncover how cells exchange signals, how those signals change in disease, and why breakdowns in communication often lead to serious health problems such as cancer, neurodegeneration, and immune disorders.
Understanding cell-to-cell communication has long been one of the biggest challenges in biology. Cells are constantly exchanging chemical signals and altering gene activity in response to their neighbors. When this process works properly, tissues grow, repair themselves, and maintain balance. When it goes wrong, disease often follows. The new tools from Duke-NUS aim to make these invisible cellular “conversations” far easier to study.
Why Cell Communication Matters So Much in Disease
Cells do not function in isolation. They live in complex neighborhoods, constantly responding to signals from nearby cells. These signals guide everything from brain activity to immune responses.
When communication fails, the consequences can be severe. In neurological conditions such as Alzheimer’s disease, disrupted signaling between neurons can lead to memory loss and cognitive decline. In cancer, tumor cells can manipulate communication pathways to send misleading signals to immune cells, allowing the tumor to grow undetected. Because of this, identifying which cells are communicating, what signals they exchange, and which genes are involved has become a central goal of modern biomedical research.
Despite its importance, studying cell communication inside real tissues has been extremely difficult. Traditional laboratory techniques struggle to capture interactions between cells in their natural environment, especially when those interactions depend on precise spatial arrangements within tissues.
The Role of Spatial Transcriptomics
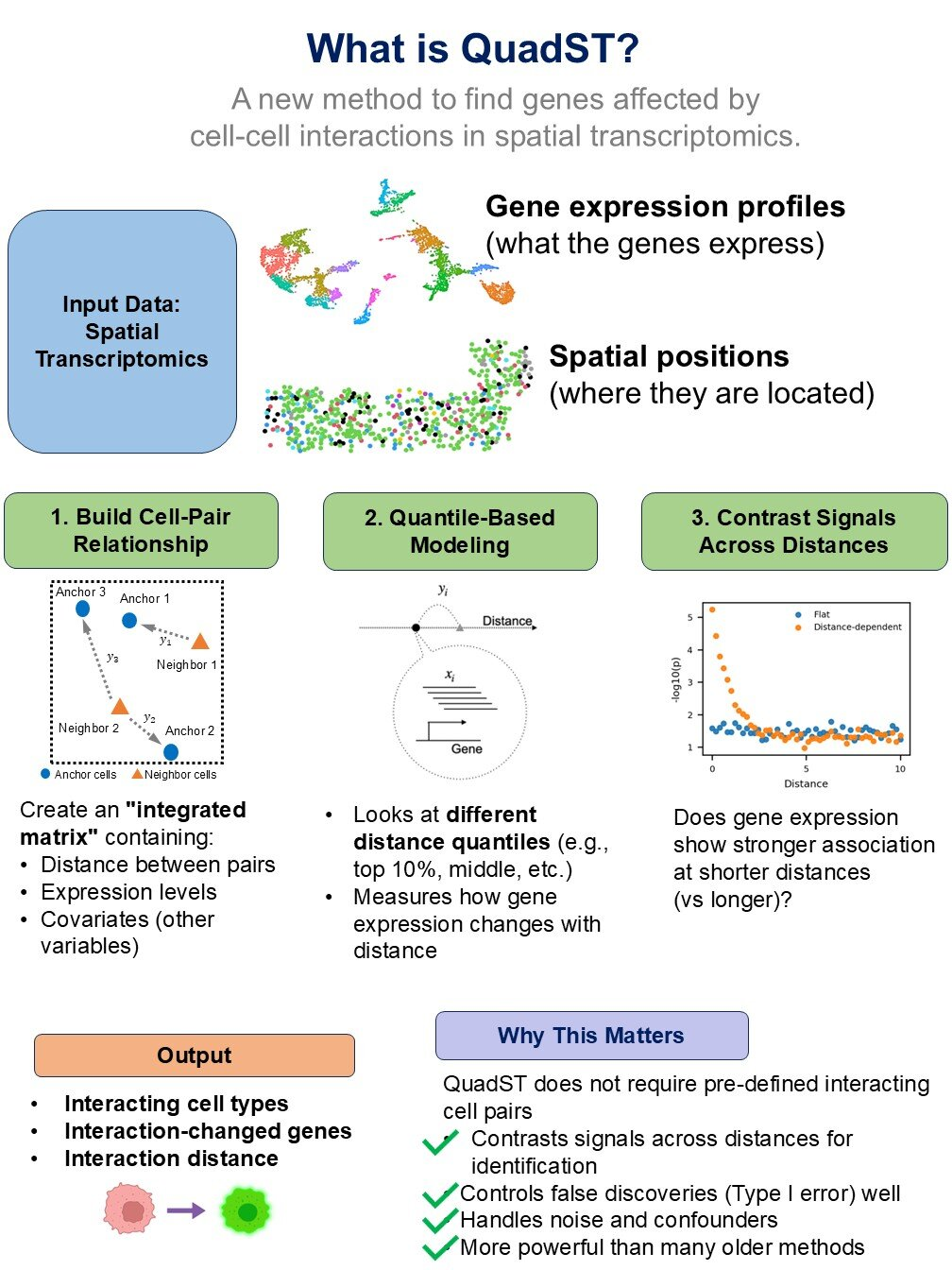
One of the most important advances in recent years has been spatial transcriptomics. This technology allows scientists to measure which genes are active in individual cells while also recording exactly where those cells are located within a tissue.
These datasets are incredibly rich, offering detailed maps of gene activity and cellular organization. However, their complexity creates a new problem. Existing computational tools often struggle to reliably analyze spatial transcriptomics data, especially when it comes to identifying genuine cell-cell communication signals.
A major challenge is the lack of ground truth. In real biological samples, researchers usually do not know which cells are truly interacting. Without that certainty, it becomes difficult to test whether computational methods are producing accurate results.
This is the gap that the Duke-NUS researchers set out to fill.
sCCIgen Creates Realistic Virtual Tissues With Known Interactions
The first tool, sCCIgen, is a high-fidelity simulator that generates realistic virtual tissues. These simulated tissues include precise information about cell locations, gene expression patterns, and communication networks between cells.
What makes sCCIgen especially valuable is that every interaction in these virtual tissues is fully known. This gives researchers a reliable benchmark for testing whether their computational methods can correctly detect cell-cell communication.
Unlike earlier simulators that relied on limited or simplified assumptions, sCCIgen is highly flexible. It can incorporate data from single-cell studies, spatial transcriptomics, and other experimental technologies. It can also model a wide range of biological scenarios, including clustered cell neighborhoods, genes whose activity changes depending on distance between cells, and gene pairs that activate only when neighboring cells interact.
Because of this flexibility, researchers can design realistic simulated experiments and evaluate how well their analytical tools capture true communication signals. In practical terms, sCCIgen acts like a training ground for computational biology, allowing scientists to refine their methods before applying them to real tissue data.
QuadST Detects Communication Signals in Real Tissues
While sCCIgen focuses on simulation, the second tool, QuadST, is designed to analyze real spatial transcriptomics datasets. Its purpose is to identify which genes and cell types are involved in communication within actual tissues.
Earlier computational approaches often relied on simplified estimates of how cells interact, which caused many important signals to be missed. QuadST improves on this by modeling how gene activity changes gradually with distance between different cell types. This approach better reflects how communication works in real biological systems.
QuadST is also built to handle noisy or incomplete datasets, which are common in large-scale biological experiments. This makes it especially useful for analyzing real-world data that may not be perfectly clean or uniform.
When applied to brain tissue, QuadST identified hundreds of genes whose activity changes when specific types of neurons interact. Many of these genes are linked to synapses, the structures that allow nerve cells to exchange signals. These findings could help researchers better understand how neuronal communication is altered in conditions such as epilepsy and neurodegenerative diseases.
The tool has also shown promise in cancer research, where it can clarify how tumor cells interact with immune cells and why some patients respond better to treatment than others.
Using sCCIgen and QuadST Together for Better Results
One of the most powerful aspects of this research is how the two tools complement each other. Scientists can first use sCCIgen to generate virtual tissues with known communication networks, allowing them to test and validate their analytical approaches. Once those methods are proven reliable, QuadST can then be applied to real tissue data to uncover genuine communication-driven gene changes.
This combined workflow increases confidence in results and reduces the risk of misinterpreting complex spatial transcriptomics data. It also makes it easier to uncover biological mechanisms that were previously hidden due to technical limitations.
How These Tools Could Impact Biomedical Research
The introduction of sCCIgen and QuadST has important implications across many fields. In cancer research, they could help identify which immune cells recognize tumors and which are being suppressed by misleading signals. In neuroscience, they could reveal how communication between neurons changes in disease. In immunology, they could clarify how immune responses are coordinated within tissues.
By pinpointing genes that change specifically due to cell-cell interactions, these tools help researchers focus on true biological signals rather than random variation. This could accelerate the discovery of new biomarkers and support the development of more precise treatments tailored to how cells communicate in different diseases.
Looking Ahead: Expanding the Virtual Cell Communication Map
The Duke-NUS research team plans to expand sCCIgen to simulate protein interactions and other molecular signaling processes, making the virtual tissues even more realistic. They also aim to use QuadST to build a reference database of genes involved in cell-cell communication across different tissues and diseases.
Such a resource would allow scientists worldwide to compare findings, validate results, and speed up discoveries in both basic research and clinical applications.
Duke-NUS Medical School continues to strengthen its reputation as a biomedical research powerhouse by combining computational innovation with translational science. With tools like sCCIgen and QuadST, researchers now have a clearer window into the complex and fascinating world of cellular communication.
Research papers:
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03762-9
https://genome.cshlp.org/content/early/2025/01/02/gr.279859.124