UNC Charlotte Scientists Fully Decode the World’s Largest Lab-Grown Bacteria-Killing Virus Using Artificial Intelligence
Researchers at the University of North Carolina at Charlotte have achieved something scientists have been chasing for over half a century — they have finally mapped the complete genome of Phage G, the largest cultivated bacteriophage known to science. With the help of artificial intelligence tools and cutting-edge computational genomics, this study provides a complete picture of a virus that has puzzled microbiologists since the late 1960s. The findings, published in npj Viruses in late 2025, could reshape how we study giant viruses and their potential to tackle antibiotic-resistant bacteria.
What Exactly Is Phage G?
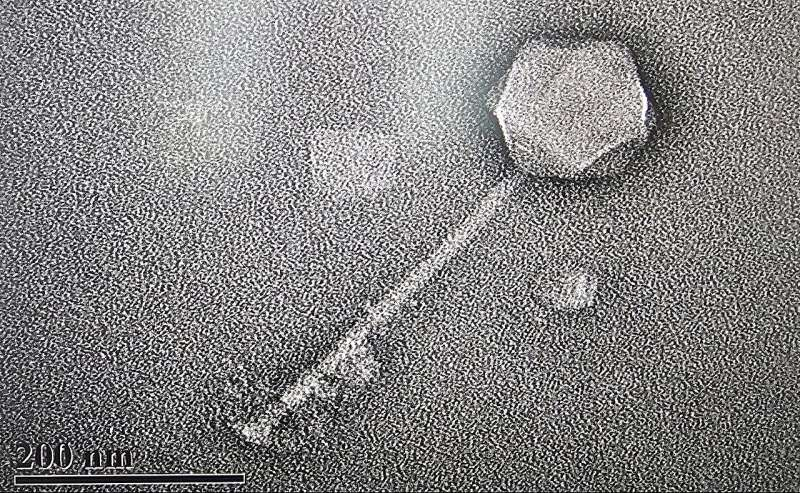
Phage G is not your average virus. It’s a bacteriophage—a virus that infects and kills bacteria instead of humans or animals. What makes Phage G extraordinary is its gigantic size. The entire virus particle stretches roughly 630 nanometers long, more than three times larger than typical phages, with a genome measuring about 499,000 base pairs.
This enormous size has fascinated scientists for decades. Phage G was first isolated in Rome in 1968 and later studied in labs across the world, but only now has its entire genome been decoded in full resolution. For comparison, most bacteriophages carry about 40,000 to 60,000 base pairs — Phage G’s genome is nearly ten times larger.
Researchers classify such oversized phages as megaphages. While megaphages are actually quite common in nature — they’ve been found in animal guts, oceans, and soil through environmental DNA sequencing — almost none have ever been grown and observed in a lab. Phage G is the one exception, making it a valuable model system for scientists to directly experiment on megaphages under controlled conditions.
The Team Behind the Discovery
The genome mapping effort was led by Andra Buchan and Stephanie Wiedman, both master’s students in Bioinformatics and Genomics at UNC Charlotte, under the guidance of Assistant Professor Richard Allen White III. The research was a collaborative project involving multiple universities: Rochester Institute of Technology, UNC Greensboro, and UT Health San Antonio.
White, who also works with UNC Charlotte’s Center for Computational Intelligence to Predict Health and Environmental Risks (CIPHER), emphasized that this project shows how AI is revolutionizing biology, especially when dealing with huge, complex datasets. Phage G’s genome isn’t just large—it’s packed with thousands of genes, many of which were previously unknown or unclassified.
Inside the Genome of a Giant Virus
Phage G’s genome is linear and non-permuted, meaning its DNA sequence is laid out end to end rather than looping into a circle. Within this enormous DNA chain, scientists identified 668 protein-coding genes. Remarkably, about two-thirds of these genes have no known function. They’re simply labeled as “hypothetical proteins.” This highlights how much remains unknown about viral evolution and diversity.
Among the recognizable genes, researchers found:
- Auxiliary metabolic genes (AMGs) that influence the host bacterium’s metabolism.
- Genes related to bacterial sporulation such as spoVK homologs.
- A suite of tRNAs, including a suppressor tRNA that helps the virus overcome genetic stop signals.
- Proteases and DNA repair enzymes, which help in managing host resources during infection.
- Defense-evading genes like anti-CBASS (Acb1) and anti-Pycsar (Apyc1) proteins, which counter bacterial immune systems.
The researchers also studied methylation patterns in Phage G’s DNA using long-read sequencing technology. They discovered a 35,000 base-pair “cryptic region” rich in methylation marks—essentially chemical tags on DNA that can regulate gene expression. This heavily methylated section contains dozens of unknown genes, making it one of the most intriguing parts of the genome.
Understanding Its Behavior and Evolution
Using AI-based prediction tools, including PhageAI and protein-structure modeling systems similar to AlphaFold2, the team predicted Phage G’s life cycle and taxonomic placement. The analysis suggests that Phage G is likely temperate—meaning it may integrate its DNA into a host’s genome and lie dormant for a while—although this has not been experimentally confirmed yet.
Taxonomically, Phage G doesn’t fit neatly into existing viral families. It sits on a distant branch within the world of megaphages but shows strong uniqueness compared to both Bacillus and Lysinibacillus-infecting viruses. The virus that scientists grow in labs today infects Lysinibacillus species, but its original natural host remains a mystery.
Interestingly, Phage G shares similarities with another megaphage discovered from moose gut DNA, although the two don’t seem to infect the same bacterial hosts. Some researchers even tell an old story about how a graduate student may have unknowingly carried Phage G into their lab decades ago—though the true origin remains unknown.
Why Cultivating a Megaphage Matters
Most megaphages discovered so far exist only in computer data. Scientists have never seen them under a microscope because they can’t grow them in lab conditions. Phage G, however, is the only one that can be cultured and physically studied. This means researchers can perform direct experiments—testing how it infects bacteria, how it replicates, and how it responds to environmental changes like temperature or pH.
The team found that Phage G loses infectivity at temperatures above 70 °C and struggles at extreme pH levels. Despite this, it remains stable enough for experimentation, giving scientists a practical model for megaphage biology.
This opens new research directions. Since bacteriophages can kill bacteria, they are seen as a promising alternative or supplement to antibiotics. As antibiotic resistance continues to grow, phages offer a natural way to target specific bacterial pathogens. Understanding how massive phages like Phage G work could lead to new therapeutic applications or genetic delivery systems in biotechnology.
The Role of Artificial Intelligence in Viral Research
The AI component of the study was crucial. Biological data is notoriously noisy and complex, and traditional bioinformatics tools struggle to interpret it when over half of the genes have no known function. By integrating machine learning models, researchers could predict which genes belong to specific viral systems, infer likely protein structures, and estimate the virus’s evolutionary relationships.
These AI-driven analyses helped classify Phage G’s genomic segments, predict its replication strategy, and identify potential interactions with its bacterial host. The researchers believe this approach will be essential as more megaphages are discovered—many of which will likely remain computational mysteries without cultivation or AI-powered interpretation.
What Makes Megaphages So Unique?
Megaphages, like Phage G, blur the line between simple viruses and complex organisms. While normal bacteriophages are minimalistic—just enough genetic material to infect and reproduce—megaphages carry hundreds of extra genes that may manipulate their hosts in intricate ways.
Some megaphages even carry their own DNA replication and transcription machinery, something rare in smaller phages. This self-sufficiency could explain why they grow slower but survive under competitive microbial conditions.
The ecological role of these giants is still being uncovered. Because they infect bacteria in diverse environments—from the human gut to marine ecosystems—they could play major roles in controlling microbial populations and influencing nutrient cycles.
What’s Next for Phage G Research?
Even with this breakthrough, many questions remain unanswered. About 66% of Phage G’s genome is still uncharacterized. Scientists don’t yet know what most of its genes actually do or why its genome contains such a large methylated region.
Understanding its true host range and environmental niche is another challenge. While researchers can grow it using Lysinibacillus bacteria, it’s unclear if that’s its natural partner in the wild.
Future work will likely focus on experimental validation—confirming predicted functions, visualizing infection dynamics, and testing whether Phage G can be used safely in biotechnology or medicine.
A Step Toward Understanding the Viral Giants
The decoding of Phage G’s genome marks a milestone in the study of bacteriophages and large DNA viruses. It’s a reminder of how much remains undiscovered in the microscopic world around us. This single virus, cultivated and studied for decades, is now revealing its secrets through the combined power of AI, advanced sequencing, and molecular biology.
As researchers continue to explore the universe of megaphages, Phage G will serve as a foundational model—an enormous, complex virus that bridges the gap between what we can culture in the lab and what we only see in DNA data.
Research Reference:
Buchan A., Wiedman S., White R. A. III et al. (2025). Unlocking the genomic repertoire of a cultivated megaphage. npj Viruses. https://doi.org/10.1038/s44298-025-00150-9