Yale Scientists Uncover Gene Pathway That Explains How Cells Break Down Their Cilia Before Division
In a recent breakthrough, researchers at Yale University have identified a previously unknown gene pathway that allows cells to disassemble their cilia — the tiny, antenna-like structures on their surfaces — before they divide. The discovery not only fills a major gap in cell biology but also sheds light on a neurological condition known as focal cortical dysplasia, a disorder often linked to seizures.
Cilia are small, hair-like projections found on the surface of many human cells. They act like microscopic antennas, sensing and transmitting signals that guide how cells grow, communicate, and respond to their environment. When a cell is preparing to divide, these cilia must be carefully broken down, or disassembled, to ensure the division process proceeds correctly. Scientists already knew a lot about how cilia are built, but how they are taken apart remained a mystery — until now.
A Closer Look at Cilia and Their Role in the Body
Before diving into the findings, it’s important to understand what cilia do. There are two main types — motile cilia, which move fluids or mucus across cell surfaces, and primary cilia, which are non-moving and act as sensory hubs for cells. The primary cilia are especially crucial for transmitting signals that regulate brain development, organ formation, and even metabolism.
Defects in cilia structure or function lead to a group of rare diseases known as ciliopathies, which can affect multiple organs, including the brain, kidneys, heart, and skeleton. Until recently, most ciliopathy research focused on how these structures form. But what happens when the body fails to remove or shorten cilia properly before cell division? That’s the question Yale scientists set out to answer.
Using CRISPR to Reverse the Usual Approach
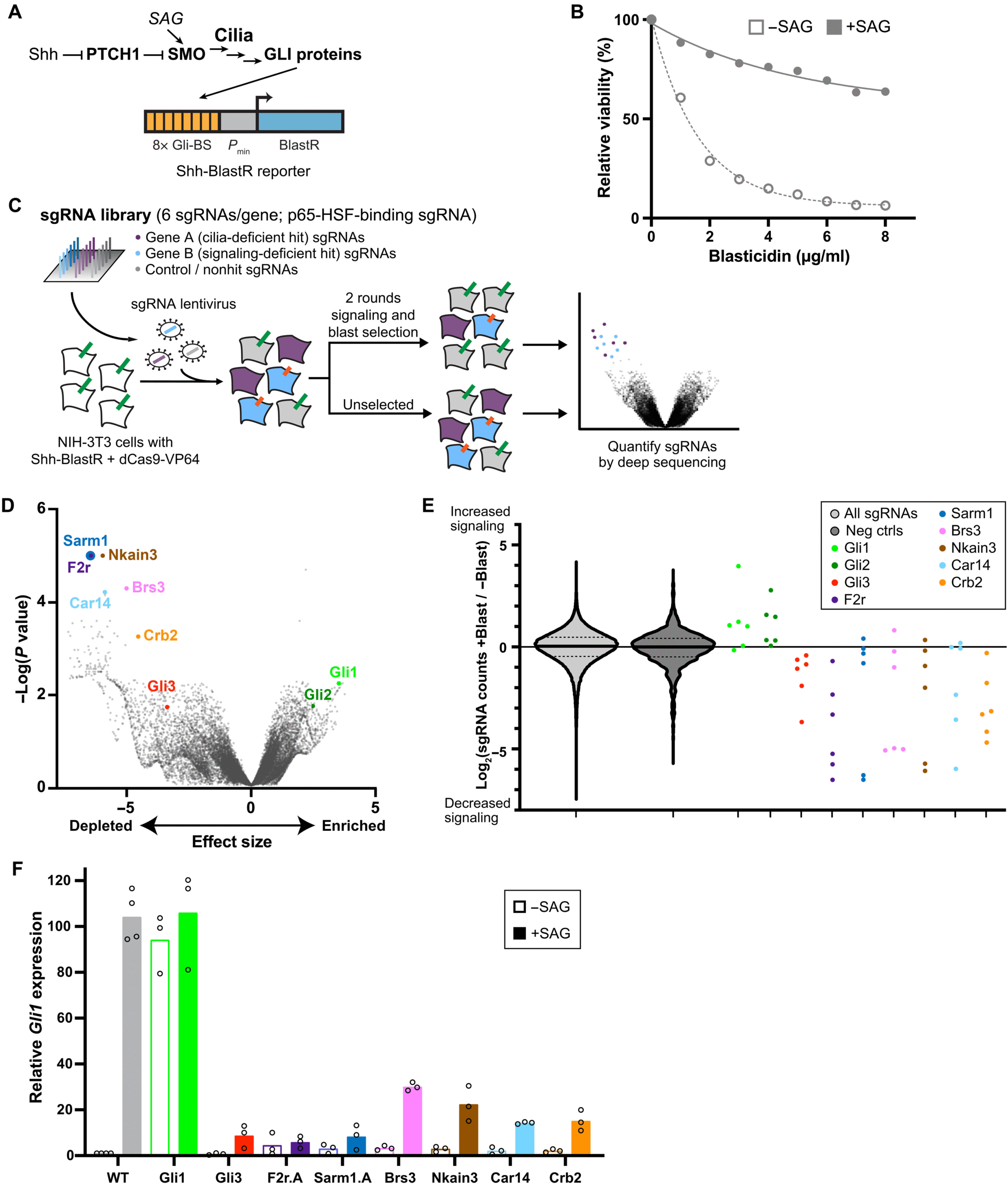
The team, led by Dr. David Breslow, took a creative approach using CRISPR activation (CRISPRa) — a variation of the popular CRISPR gene-editing technology. Typically, CRISPR is used to turn off genes to see what functions they control. But in this case, the researchers did the opposite: they used CRISPRa to activate every gene in the genome to see which ones caused cells to lose their cilia.
The experiment was done in NIH-3T3 cells, a type of mouse fibroblast cell commonly used in laboratory studies. By turning up the activity of thousands of genes, the researchers could see which ones triggered the breakdown of cilia. This large-scale screen, which would have been impossible just a few years ago, revealed something remarkable — two genes stood out for their strong effects on cilia disassembly: F2R and SARM1.
The Discovery of the F2R–SARM1 Pathway
The gene F2R, also known as protease-activated receptor 1 (PAR1), is part of a family of G protein-coupled receptors (GPCRs) that are well known for their role in cell signaling. When the team increased F2R activity, cells began rapidly losing their cilia. They found that this process could be blocked by a drug called vorapaxar, an F2R inhibitor, confirming the gene’s involvement in cilia breakdown.
The second key player, SARM1, is an enzyme known for its role in nerve damage and axon degeneration. SARM1 contains an NADase domain, which breaks down NAD⁺ molecules — a key player in cellular metabolism — into nicotinamide (NAM) and cyclic ADP-ribose (cADPR). When the Yale team overactivated SARM1, cilia also disassembled. However, using a SARM1 inhibitor called DSRM-3716 prevented this from happening, suggesting that its enzymatic activity directly contributes to the breakdown process.
The researchers pieced together the pathway as follows: activation of F2R triggers G protein signaling, which leads to calcium release near the cell’s centrosome via ryanodine receptors. This calcium surge activates RhoA, a protein that coordinates structural changes in the cell, which in turn activates SARM1. The end result? The disassembly of cilia.
Linking the Pathway to a Neurological Disorder
The most surprising part of the discovery came when the scientists compared their findings to existing data on focal cortical dysplasia (FCD), a rare neurological condition. FCD causes abnormal brain development and is one of the leading causes of drug-resistant epilepsy in children.
When the team examined genetic studies of FCD patients, they noticed something striking — mutations in several genes that overlapped with the ones in their newly identified cilia disassembly pathway. These included the same F2R, RhoA, and SARM1 genes they had studied in the lab.
To test whether these mutations affected cilia, the researchers expressed patient-derived mutant versions of these genes in cells. The results confirmed their suspicion: the mutations caused loss of cilia, mimicking the same disassembly effect they saw with overactivation of the genes. Interestingly, when they treated these cells with SARM1 inhibitors, the cilia grew back, suggesting that blocking excessive disassembly could be a potential therapeutic strategy.
This connection hints that the brains of FCD patients may suffer from abnormal cilia loss in developing neurons, disrupting how those cells communicate and organize themselves during brain formation.
Implications for Medicine and Beyond
Understanding this cilia disassembly pathway could reshape how scientists think about neurological diseases — and possibly even cancer. In both cases, abnormal cilia loss has been observed, but the molecular reasons were unclear. This study shows that too much cilia disassembly can be just as damaging as too little cilia formation.
The discovery also opens the door to targeted therapies. Because F2R and SARM1 are both druggable proteins (with existing inhibitors already used or tested in other conditions), researchers can now explore whether controlling this pathway could restore normal cilia function in diseases caused by their overactivity.
It’s worth noting that SARM1 is already a hot topic in neuroscience. The same enzyme is known for promoting axon degeneration — the breakdown of nerve fibers — after injury. Pharmaceutical companies have been investigating SARM1 inhibitors as treatments for neurodegenerative diseases. Now, this study adds another layer: blocking SARM1 could also help preserve cellular cilia, potentially improving brain cell health.
Why Cilia Disassembly Matters
Most people think of cilia as permanent structures, but in reality, they are dynamic — constantly assembling and disassembling depending on what the cell is doing. During cell division, a cell needs to remove its cilium so it can properly duplicate its DNA and divide into two new cells. After division, the cilium can reform. If this process doesn’t happen correctly, it can lead to abnormal cell growth, developmental defects, or signaling imbalances.
For example, in cancer cells, researchers have noticed that primary cilia are often missing, which could alter key signaling pathways that normally control growth. Similarly, in neurological disorders like FCD, losing cilia prematurely may disturb how neurons connect and communicate.
This new study gives scientists a detailed look at the molecular machinery driving that disassembly process, allowing future research to test whether manipulating it can treat disease.
The Road Ahead
The Yale team plans to continue exploring how cilia disassembly works in neurons and how it relates to brain development. They are particularly interested in how calcium signaling and metabolic enzymes like SARM1 coordinate the process, and whether restoring cilia could correct developmental issues in disease models.
As Dr. Breslow’s group suggested, understanding this balance between cilia assembly and disassembly could lead to better strategies not just for treating FCD but also for other diseases where cilia play a role — including kidney disease, skeletal disorders, and certain cancers.
The research also highlights the power of CRISPRa screening — a technology that can systematically increase gene activity to uncover hidden biological pathways. It’s the reverse of traditional CRISPR knockout methods, and this study shows how effective it can be in revealing new biology that might otherwise remain unseen.
Reference
Research Paper: A CRISPR activation screen reveals a cilia disassembly pathway mutated in focal cortical dysplasia – Science Advances (2025)