Genome-Wide Gene Study in Sorghum Uncovers Key Regulators That Could Boost Stem Biomass for Bioenergy
A new study has mapped how genes behave across the different organs and growth stages of sorghum, revealing the inner workings that make its stems such a rich source of biomass. Scientists at the University of Illinois at Urbana-Champaign, along with collaborators from other research centers, have taken a deep dive into the genome-wide expression patterns of this resilient crop, identifying crucial stem-specific genes and transcription factors that could hold the key to improving its performance as a bioenergy feedstock.
Why Sorghum Matters
Sorghum bicolor is a heat- and drought-tolerant crop that thrives where other grains might fail. It grows deep roots, performs well on marginal lands, and has long been valued for its versatility — serving as food, animal feed, and increasingly as a renewable resource for biofuels, bioplastics, and biopolymers. What makes it particularly promising for bioenergy is its sturdy stem, which contains the majority of its biomass — including both lignocellulosic material and soluble sugars that can be converted into energy or sustainable materials.
Despite major advances in sorghum genomics and transcriptomics, one important question remained largely unanswered: Which genes are truly responsible for building and maintaining those thick, energy-rich stems? The new study provides one of the most comprehensive answers yet.
What the Researchers Did
The research team performed a reanalysis of existing RNA-sequencing (RNA-seq) data, studying how genes are turned on and off in different parts of the sorghum plant — including stems, leaves, roots, and seeds — across five major developmental stages: juvenile, vegetative, floral initiation, anthesis (flowering), and grain maturity.
Their goal was to identify organ-preferential genes, especially those with stem-focused activity, and to understand how these genes change as the plant grows. By doing so, they hoped to find genetic “switches” that could be used to enhance sorghum’s biomass through precise genetic engineering.
Using computational tools such as Weighted Gene Co-expression Network Analysis (WGCNA), the scientists grouped genes into clusters (or modules) based on similar expression patterns. They then measured how strongly each gene was tied to specific organs using a “Tau” metric — a common measure of tissue specificity.
Interestingly, they found that stem-specific genes are fewer than those specific to leaves or roots, but they tend to be highly specialized and play central regulatory roles. This makes them especially valuable targets for crop improvement.
Stage-by-Stage Stem Biology
The team’s analysis revealed that the functions of stem genes evolve throughout the plant’s life. Early in growth (juvenile and vegetative stages), stem genes are mostly involved in cell division and tissue development. Later, during flowering and grain formation, the same genes shift toward functions like cell wall thickening, lignin biosynthesis, and energy storage.
This stage-dependent pattern shows that the sorghum stem is not just a static structure; it’s a dynamic organ, constantly reprogramming its internal processes as the plant matures.
Identifying Stem-Specific Genes Across Genotypes
To make sure these findings weren’t limited to one variety, the researchers tested their gene sets across multiple sorghum genotypes, including grain, sweet, and biomass types. Many genes retained the same stem-preferential expression across these varieties, suggesting a universal stem identity program that runs consistently in different sorghum backgrounds.
These core stem genes are of particular interest because they can be used in diverse breeding and genetic engineering programs without worrying about genotype limitations.
Promoter Motifs: The Switches That Control Expression
Beyond identifying which genes are active, the study also examined promoter regions — the stretches of DNA that control when and where a gene is expressed. The researchers analyzed about 1.5 kilobases of upstream DNA sequences for each stem-specific gene to find promoter motifs (short DNA patterns where transcription factors bind).
They discovered that each developmental stage had its own unique set of enriched motifs, indicating that the molecular switches controlling stem expression change as the plant develops. For instance, early-stage motifs were linked to genes involved in growth and differentiation, while late-stage motifs were associated with hormone signaling and secondary cell wall formation.
Two motifs stood out: MA0965.1, common to juvenile and vegetative stages, and MA0937.1, found across anthesis and grain maturity stages. These motifs could become valuable tools for designing stage-specific promoters that activate desired genes only at particular points in the plant’s life.
The Big Discovery: SbTALE03 and SbTALE04
Among hundreds of genes analyzed, two transcription factors emerged as major players: SbTALE03 (Sobic.001G106000) and SbTALE04 (Sobic.001G106200). Both belong to the TALE (Three Amino-Acid Loop Extension) homeodomain family — known for controlling plant growth and development — and both appear to function as “hub regulators” within the stem’s gene network.
What makes them remarkable is that they were identified as hub genes across all developmental stages, suggesting they’re central to maintaining the identity and growth of the sorghum stem from seedling to maturity.
Further analysis showed that these genes likely regulate a wide variety of targets, including those related to cell wall biosynthesis, hormone pathways, and epigenetic control. Early in development, SbTALE03 and SbTALE04 connect with genes that maintain meristematic (undifferentiated) cell activity. Later on, they interact with genes tied to hormone biosynthesis, lipid signaling, and histone methylation — reflecting a complete reprogramming of the stem’s regulatory system as the plant matures.
Experimental Validation
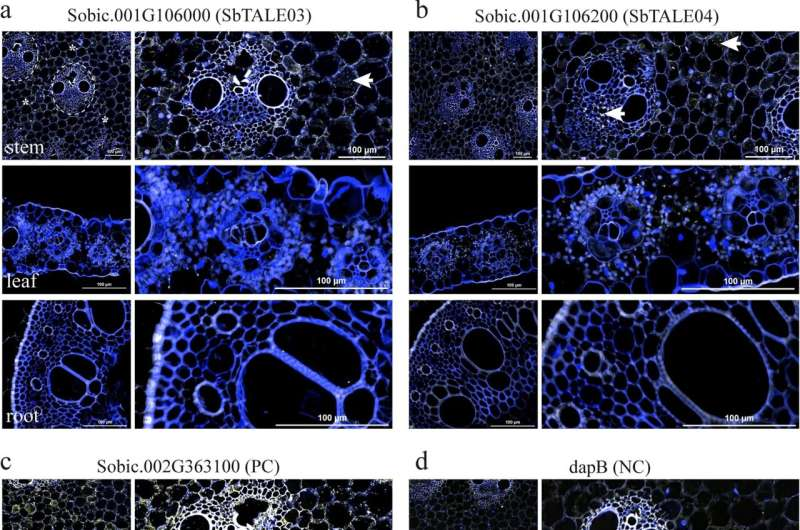
To confirm their computational predictions, the team used RNAscope in situ hybridization (ISH), a technique that visualizes where specific RNA molecules appear in plant tissues. The tests showed that SbTALE03 and SbTALE04 had strong expression in stems — particularly in pith parenchyma and vascular tissues — but very low or undetectable expression in leaves or roots.
Quantitatively, the expression levels were 5.7 to 26.6 times higher in stems than in other organs. This clear spatial pattern supports their designation as stem-specific transcription factors.
The researchers went a step further and conducted electrophoretic mobility shift assays (EMSA) to see if SbTALE03 could physically bind to DNA sequences in the promoters of its predicted target genes. SbTALE03 successfully bound to six out of seven promoter sequences tested, providing strong biochemical evidence that it indeed acts as a regulatory hub. SbTALE04 could not be tested due to difficulties in producing a stable protein sample.
Why It Matters for Bioenergy
For those working to turn sorghum into a better biofuel or bioproduct feedstock, these findings are exciting. The identified stem-specific genes and promoters could be used to engineer plants with higher biomass, improved cell wall composition, or optimized sugar content — all without affecting other plant parts.
This is critical because using organ-specific promoters avoids the negative side effects often seen when genes are turned on everywhere in the plant. For example, modifying lignin biosynthesis globally can weaken leaves or roots, but if the same modification is limited to the stem, overall plant health remains intact while the desired trait is achieved.
Broader Implications and Future Directions
The discovery of SbTALE03 and SbTALE04 as universal stem regulators opens the door for targeted manipulation of sorghum and possibly other C4 grasses like maize and sugarcane. Interestingly, these two genes are tandem duplicates and are homologous to known maize genes involved in stem development, hinting that this regulatory network might be conserved across related species.
Going forward, scientists could explore how modifying these transcription factors — or using their promoters to drive new genes — might influence plant growth, sugar accumulation, or stress resilience. The dataset also provides a foundation for identifying other hub genes involved in the same networks.
The Bigger Picture of Sorghum Research
Sorghum has already proven itself as a climate-resilient crop, capable of producing good yields under water-limited conditions. Combining that resilience with new genetic tools to boost stem biomass could make it a cornerstone of future bioenergy and sustainable agriculture strategies.
This study not only enhances our understanding of how sorghum builds its stems but also highlights the power of modern genomics in unlocking nature’s blueprints for more efficient, sustainable crops.
Research Reference:
Jie Fu et al. (2025). Stage-resolved gene regulatory network analysis reveals developmental reprogramming and genes with robust stem-preferred expression in sorghum. BMC Plant Biology. https://doi.org/10.1186/s12870-025-07303-1