A Deep-Learning Model Can Now Predict How Fruit Flies Form Cell by Cell During Early Development
Early development is one of the most complex and fascinating processes in biology. From a seemingly uniform cluster of cells, entire tissues and organs begin to take shape through constant motion, division, folding, and reorganization. Scientists have studied this process for decades, but predicting exactly how thousands of individual cells behave over time has remained a major challenge. Now, researchers at MIT have developed a powerful deep-learning model that can do exactly that—predict how fruit fly embryos form, cell by cell and minute by minute, during the earliest stage of life.
The research, published in the journal Nature Methods, introduces a new computational framework that allows scientists to forecast how individual cells will move, divide, fold, and interact with their neighbors as an embryo develops. While the study focuses on fruit flies, the implications extend far beyond insects, opening new possibilities for understanding development, disease, and tissue formation in more complex organisms.
Predicting Development One Cell at a Time
During early development, tissues and organs do not simply grow larger. Instead, thousands of cells shift position, split into new cells, attach to neighbors, detach from others, and fold into complex shapes. These processes are especially intense during gastrulation, the first major developmental phase in many animals.
In fruit flies, gastrulation occurs during the first hour of development. At this stage, the embryo starts as a roughly smooth, ellipsoid shape made up of about 5,000 cells. Over the course of just 60 minutes, this smooth surface transforms dramatically as folds appear and cells rearrange to establish the body’s basic structure.
The MIT team developed a deep-learning model capable of predicting how each of these 5,000 cells will behave at each moment during this crucial hour. When tested, the model achieved an impressive 90 percent accuracy, correctly forecasting whether individual cells would fold, divide, shift position, or maintain contact with neighboring cells.
A New Way to Model Living Tissues
Traditionally, scientists have used two main approaches to model embryonic development. One treats cells as points in space—known as a point cloud model—where each point moves over time. The other represents cells as soft, bubble-like shapes that press and slide against each other, often referred to as a foam model.
Each approach captures part of the story, but neither fully represents the complexity of living tissues. Instead of choosing between the two, the MIT researchers combined them.
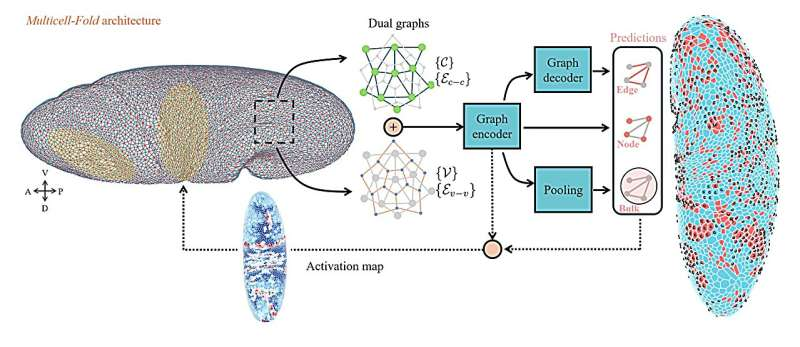
At the core of their new system is a dual-graph representation. In this framework, cells are modeled both as moving points and as connected shapes that share edges with neighboring cells. This dual structure allows the model to capture detailed geometric and topological information, such as cell position, cell-to-cell contact, nuclear location, and changes in shape or connectivity over time.
By representing the embryo as a dynamic graph, the model can track how local interactions between neighboring cells lead to large-scale tissue folding and organization.
Training the Model With Rare, High-Quality Data
One of the most remarkable aspects of this research is the quality of the data used to train the model. The researchers worked with collaborators at the University of Michigan, who provided extremely detailed 3D videos of fruit fly embryos undergoing gastrulation.
These videos capture the entire embryo at single-cell resolution, with submicron spatial detail and a fast frame rate. Each cell’s edges and nucleus are labeled, making it possible to track individual cells accurately as they move and change shape.
Such datasets are exceptionally rare, both because of the technical difficulty of imaging entire embryos at this resolution and the challenge of labeling thousands of cells over time. Using data from three embryo videos, the researchers trained the model to learn how cells typically behave. They then tested the model on a fourth, previously unseen video.
The result was a system that could not only predict what would happen to each cell, but also when it would happen—down to differences of just a minute.
Why Gastrulation Matters So Much
Gastrulation is one of the most important stages of development in animals. It is during this phase that cells reorganize to form the basic layers and structures that later give rise to organs such as the gut, muscles, and nervous system.
Errors during gastrulation can have serious consequences, leading to developmental defects or disease. By accurately modeling this stage, scientists can begin to understand how local cell interactions scale up into global tissue patterns.
The new deep-learning approach provides a way to study these processes quantitatively, rather than relying solely on visual observation or simplified theoretical models.
Extending the Model Beyond Fruit Flies
Although fruit flies are a classic model organism in biology, the researchers see this work as a stepping stone toward understanding development in other species. In principle, the same approach could be applied to organisms such as zebrafish, mice, and eventually even human tissues.
From a computational perspective, the model is already capable of handling more complex systems. The main limitation is the availability of equally high-quality imaging data. Without detailed, labeled videos of developing tissues, even the most advanced models cannot make accurate predictions.
Still, as imaging technologies continue to improve, datasets like the ones used in this study may become more common.
Potential Applications in Disease Research
One of the most exciting long-term possibilities of this work lies in disease research. Many diseases are associated with abnormal tissue structure, but scientists often do not know when or how those abnormalities first emerge.
For example, lung tissue in people with asthma looks noticeably different from healthy lung tissue. The early developmental steps that lead to these differences are not well understood. By comparing predicted cell dynamics in healthy versus disease-prone tissues, researchers may be able to identify subtle changes that occur long before symptoms appear.
The same approach could potentially be applied to diseases like cancer, where changes in cell behavior and organization play a critical role.
Why This Matters for Biology and AI
This research represents a growing trend at the intersection of biology, physics, and artificial intelligence. Rather than using AI only to classify images or analyze static data, this work shows how deep learning can be used to model dynamic, physical processes in living systems.
By embedding biological structure directly into the model—through geometric and graph-based representations—the researchers created a system that respects the underlying rules of tissue organization. This makes the predictions not only accurate, but also biologically meaningful.
Looking Ahead
The MIT team believes that their model is largely ready for broader application. The real bottleneck now is data. As better imaging techniques emerge and more high-quality developmental datasets become available, models like this could become essential tools for developmental biology, regenerative medicine, and disease research.
For now, this work marks a significant step forward in understanding how life organizes itself from the very beginning—one cell at a time.
Research paper:
MultiCell: geometric learning in multicellular development, Nature Methods (2025)
https://doi.org/10.1038/s41592-025-02983-x