AI Is Mapping Hidden Gene Redundancies to Help Scientists Build Stronger Crops
Artificial intelligence is steadily making its way into plant science, and a new study from researchers at Cold Spring Harbor Laboratory shows just how powerful it can be. Scientists have developed an AI-driven system that maps gene redundancies in plants and predicts how genetic changes might affect important traits like growth and development. The work offers what researchers describe as a practical roadmap for future crop improvement, especially as climate change places increasing pressure on global agriculture.
Why Gene Redundancy Has Been a Major Roadblock
One of the biggest challenges in crop genetics is that plants rarely rely on a single gene to control an important trait. Over millions of years of evolution, plant genomes have undergone repeated gene duplication events. When a gene duplicates, both copies can survive and continue to function. This often leads to redundancy, where multiple related genes can compensate for one another.
For plant breeders and genetic engineers, this creates a serious problem. Editing or mutating one gene may do nothing at all if other genes quietly take over its role. As a result, researchers often struggle to identify which genes truly matter when trying to improve traits such as yield, plant size, or stress resistance.
The Cold Spring Harbor team set out to address this issue directly by asking a fundamental question: after a gene duplicates, how do the copies evolve, and which ones remain functionally overlapping?
Tracking a Gene Family Across 140 Million Years
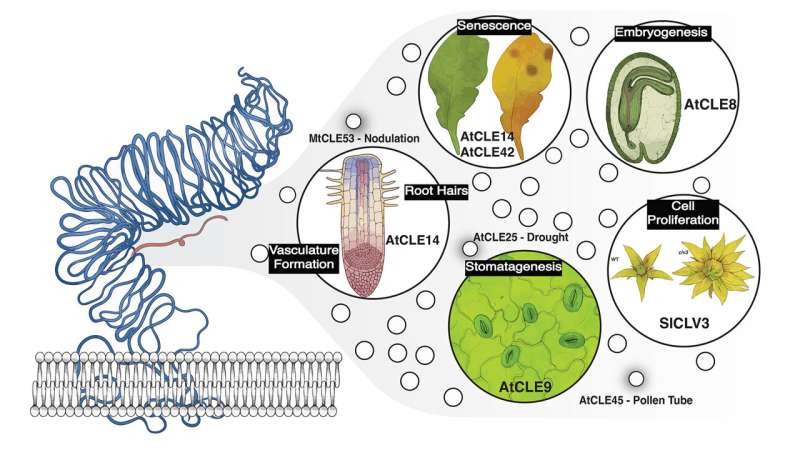
To answer that question, the researchers focused on a specific group of genes known as the CLE gene family. CLE genes produce short signaling peptides that play key roles in plant development, including cell division, organ formation, and growth regulation. These peptides are found in virtually all flowering plants, making them an ideal system for studying evolutionary patterns.
However, CLE genes are notoriously difficult to analyze. Their peptides are very short, they evolve rapidly, and they often exist in large, redundant clusters. Traditional genetic methods struggle to make sense of them.
Using modern AI techniques, the team conducted a pan-angiosperm analysis, examining CLE genes across roughly 1,000 plant species. This allowed them to trace how the gene family has changed over approximately 140 million years of evolution.
In the process, the AI system identified thousands of CLE genes that had not been previously recognized, significantly expanding scientific understanding of this gene family.
How AI Was Used to Predict Redundancy
Once the researchers assembled this massive dataset, they trained computational models to detect patterns associated with redundancy. The AI looked for similarities in two main areas:
- Peptide sequences, which determine the biochemical activity of the gene’s product
- Promoter regions, which control when and where a gene is expressed
The models flagged genes that were likely to be redundant, meaning that editing one alone would probably have little effect. Interestingly, the analysis revealed that redundancy often depends more on shared promoter features than on identical peptide sequences. In other words, genes can produce different peptides yet still act redundantly if they are switched on in the same tissues at the same time.
This insight helps explain why many past genetic experiments failed to produce visible changes: researchers were targeting genes without realizing that multiple backups were active in parallel.
Putting the Predictions to the Test With CRISPR
To confirm that the AI’s predictions reflected biological reality, the team moved from computation to experimentation. They used CRISPR gene editing to knock out predicted redundant CLE genes in tomato plants, a widely used model crop.
When individual genes were edited one at a time, the plants showed little or no change, exactly as the model predicted. But when the researchers simultaneously knocked out multiple redundant genes, the plants displayed clear and dramatic developmental changes.
In one experiment, as many as 10 CLE genes were targeted at once. This large-scale, multi-gene editing demonstrated that redundancy had been masking important genetic functions all along.
Predicting the Impact of Genetic Changes
Beyond identifying redundant genes, the AI system was also trained to predict the outcomes of specific mutations. For each gene or gene group, the model could estimate whether altering it would likely have a positive, negative, or neutral effect on plant traits.
This predictive ability is especially valuable for crop improvement. Instead of relying on trial and error, breeders and genetic engineers can now prioritize edits that are more likely to produce useful results while avoiding changes that might harm the plant.
Why This Matters for Crop Improvement
As global temperatures rise and weather patterns become more unpredictable, agriculture faces growing challenges. Crops must be more resilient, efficient, and adaptable than ever before. Genetic improvement remains one of the most powerful tools available, but gene redundancy has long slowed progress.
The approach developed by the Cold Spring Harbor team offers a way forward. By revealing hidden layers of redundancy, the AI system helps scientists see which genes truly matter and how they interact. This could accelerate the development of crops with improved yield, better stress tolerance, and more predictable growth characteristics.
Importantly, the researchers emphasize that this framework is not limited to CLE genes. In principle, the same method can be applied to any gene family in plants, opening the door to broad improvements across many species.
A Broader Look at AI in Plant Genetics
This study fits into a larger trend of using AI and machine learning in plant biology. Modern plant genomes are vast and complex, and traditional analysis methods struggle to keep up. AI excels at detecting patterns across large datasets, making it well suited for evolutionary studies and gene function prediction.
In recent years, AI has been applied to areas such as genomic selection, trait prediction, and phenotyping. What makes this work stand out is its focus on evolutionary history and functional redundancy, two areas that are critical for understanding why certain genetic interventions succeed or fail.
Understanding CLE Peptides and Their Roles
CLE peptides themselves are fascinating molecules. Despite their small size, they act as powerful signaling agents, coordinating how cells divide and differentiate within plant tissues. They influence processes ranging from stem cell maintenance to root architecture.
Because CLE genes often overlap in function, their importance has been underestimated. This new work helps clarify their roles and highlights how subtle genetic networks shape plant development.
What Comes Next
The researchers describe their system as a starting point rather than a finished solution. As more plant genomes become available and AI models continue to improve, predictions about gene function and redundancy will become even more accurate.
For plant breeders, this means better tools for making informed decisions. For scientists, it offers deeper insight into how genomes evolve and adapt. And for agriculture as a whole, it represents a step toward more efficient, targeted, and sustainable crop improvement.
Research paper:
https://academic.oup.com/mbe/article/42/1/msaf294/7590610





