Cellarity Unveils a Groundbreaking AI Framework to Discover Medicines That Correct Diseased Cell States
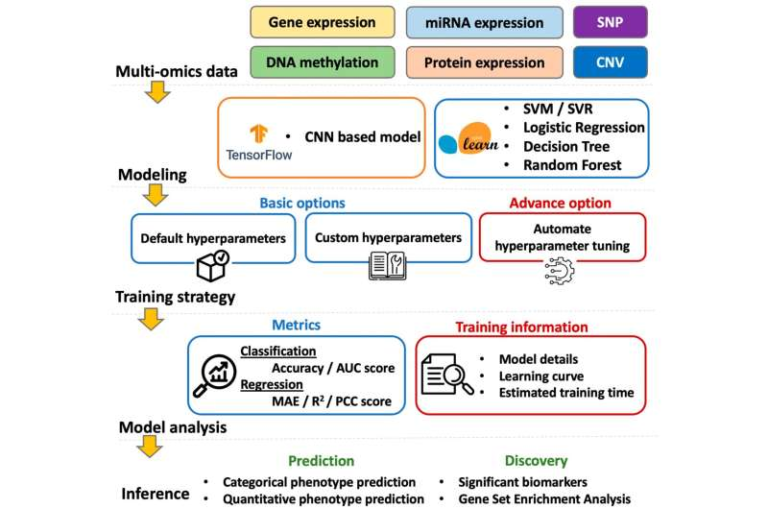
A major step forward in biotechnology has just been announced by Cellarity, a Massachusetts-based biotech company focused on developing cell state-correcting medicines. The company recently published a study in Science (October 2025) describing a new AI-driven, active learning framework that integrates single-cell transcriptomics and multi-omics data to accelerate the discovery of drugs that can restore normal cellular function in complex diseases.
The publication marks a significant milestone for the industry, outlining a blueprint for integrating artificial intelligence into drug discovery in a way that directly links chemistry to disease biology. The study’s results show that this approach can make drug discovery not only faster but also more accurate—by focusing on the cell’s overall state, instead of just one genetic target.
The Core of Cellarity’s Approach
Traditional drug discovery usually revolves around finding a single molecular target—often a protein—and designing a compound to inhibit or activate it. While that strategy has produced many successful drugs, it struggles with diseases caused by complex cellular dysfunction rather than a single faulty gene.
Cellarity’s approach is different. It focuses on the entire cell state, meaning the complete set of molecular processes, pathways, and interactions that define how a cell behaves. By analyzing how these states change during disease and under chemical perturbation, the company aims to identify drugs that restore diseased cells to a healthy condition.
Their discovery platform combines high-dimensional single-cell transcriptomics with AI models that learn to predict which compounds can drive those restorative changes. Essentially, the system maps the biology of disease at the cellular level, then matches it with the chemistry most likely to correct it.
The Active Learning Framework
At the heart of the new publication is a lab-in-the-loop active learning framework—a closed feedback system where experiments and AI models constantly improve each other. Here’s how it works:
- Large-scale transcriptomic data are collected at the single-cell level after cells are exposed to various chemical compounds.
- Deep learning models analyze this data to predict which compounds can move diseased cell states toward healthy ones.
- Predictions guide the next round of lab experiments. The new data then refine the model further, making each iteration smarter and more accurate.
This continuous refinement loop led to an impressive 13- to 17-fold improvement in recovering active compounds compared to standard phenotypic drug screening. When the researchers added an extra refinement step—called signature optimization—the improvement doubled again, proving that data-driven iteration can dramatically boost discovery efficiency.
The study demonstrated these gains across multiple disease-relevant contexts, particularly within hematological cell systems, which are key to understanding disorders like anemia and leukemia.
Beyond Target-Based Discovery
One of the biggest challenges in modern drug discovery is that most diseases are not caused by a single mutation or pathway. Instead, they emerge from network-level dysfunctions involving many interacting biological systems.
Cellarity’s scientists argue that understanding and correcting cell states provides a broader, more realistic view of disease biology. Their approach captures how genes, proteins, and epigenetic factors collectively determine cellular health. This holistic understanding could help overcome the limitations of target-centric drug design, which often fails in complex diseases such as neurodegenerative disorders, metabolic syndromes, or immune dysregulation.
By leveraging AI-based modeling, the company’s platform connects chemistry to cellular biology in a way that traditional pipelines simply cannot. It helps identify multi-target or network-acting compounds, also known as polypharmacological agents, which may be more effective at treating multifactorial diseases.
Datasets Released for the Scientific Community
In conjunction with the publication, Cellarity released several large-scale single-cell datasets to encourage collaboration and independent benchmarking. These open resources are designed to help scientists and developers worldwide explore the dynamics of cell states under chemical perturbation.
- Perturbational Transcriptomic Dataset:
- Includes over 1,700 samples and about 1.26 million single cells.
- Enables mapping of drug responses across multiple cell types.
- Can be used to benchmark new algorithms for perturbation prediction and cross-cell-type response analysis.
- Single-Cell Multi-omic Hematopoiesis Atlas:
- Integrates transcriptomic, surface receptor, and chromatin accessibility data.
- Provides a multi-layered view of blood cell formation (hematopoiesis).
- Used in the publication to identify precise signatures of megakaryopoiesis (platelet-forming cells) and erythropoiesis (red blood cell formation).
- Megakaryocyte Differentiation Timeline Dataset:
- Captures the process of megakaryocyte (Mk) maturation under chemical perturbation.
- Helps track time-resolved drug effects and understand how interventions shift developmental trajectories.
These datasets are publicly accessible, allowing scientists to perform cross-platform analyses, train new machine learning models, and potentially uncover previously unknown aspects of cellular regulation.
The First Drug Candidate from the Platform
The company’s first compound developed through this platform, CLY-124, is currently in a Phase I clinical trial for sickle cell disease. While early-stage trials focus primarily on safety, this marks an important step in proving that Cellarity’s theoretical framework can lead to real, testable drugs.
If successful, it will not only validate the company’s AI-integrated approach but also demonstrate that cell-state correction can be turned into viable therapeutic strategies for complex diseases.
Why This Matters
Drug discovery has long struggled with declining success rates. Despite enormous investments, the probability of bringing a new drug to market remains extremely low. One reason is that conventional pipelines rely heavily on simplified models of disease that don’t reflect the intricate biological reality inside living cells.
By contrast, Cellarity’s platform embraces complexity. Its models learn from massive, high-resolution datasets and use AI-driven prediction loops to pinpoint compounds that meaningfully shift disease biology. This could make drug discovery more predictive and less reliant on chance.
If broadly adopted, such frameworks might shorten development timelines, reduce failure rates, and expand the universe of treatable diseases—particularly in areas where traditional approaches have stagnated.
Understanding Key Concepts
To appreciate the significance of Cellarity’s work, it helps to unpack some of the underlying concepts.
Transcriptomics: This refers to the study of all RNA molecules expressed by cells, providing a snapshot of which genes are active and how they respond to stimuli. In this study, transcriptomics at single-cell resolution allowed scientists to see how individual cells—rather than bulk populations—react to compounds.
Multi-omics: Beyond RNA, multi-omics integrates data from several biological layers: genomics, proteomics, metabolomics, and epigenomics. Combining these layers gives a more comprehensive view of cellular function and regulation.
Active Learning: A machine learning approach where the algorithm doesn’t just passively analyze data—it actively decides what data it needs next to improve. In this case, the AI system suggested which chemical perturbations to test, creating a continuous loop between computational prediction and lab experimentation.
Polypharmacology: Many effective drugs act on multiple targets, not just one. By capturing whole-cell states, Cellarity’s platform naturally identifies such multi-target compounds, which may be more robust in treating complex diseases.
Collaboration and Broader Impact
The research represents a collaboration between Cellarity, the Broad Institute of MIT and Harvard, MIT’s Department of Biological Engineering, and Helmholtz Munich. Notably, the team includes Jim Collins, a pioneer in synthetic biology, and Alex Shalek, known for his expertise in single-cell analysis.
This partnership demonstrates the growing convergence of AI, biology, and data science in drug discovery. It also sets an example for how biotech companies can promote transparency and community progress by sharing data openly.
Looking Ahead
While still in early stages, this framework has broad potential. If similar success is achieved across different tissues and diseases, AI-guided cell-state correction could transform how we think about treating multifactorial disorders—from fibrosis to autoimmune diseases to neurodegeneration.
The field will be watching closely as the CLY-124 trial unfolds and as researchers worldwide begin to analyze the newly released datasets. Whether this truly ushers in a new era of AI-enabled drug discovery will depend on how these findings translate into clinical outcomes.
Research Reference:
Benjamin DeMeo et al., “Active learning framework leveraging transcriptomics identifies modulators of disease phenotypes,” Science, 2025. DOI: 10.1126/science.adi8577