Decoded Rules of microRNA Strand Selection Reveal Conserved and Programmable Features
MicroRNAs are tiny molecules, but their influence on biology is enormous. These short, non-coding RNA sequences play a central role in regulating gene expression, helping cells decide which genes should be turned on, turned down, or silenced altogether. In 2025, the discovery of microRNAs was formally recognized with the Nobel Prize in Physiology or Medicine, highlighting just how foundational they are to modern biology and medicine.
Despite decades of research, one surprisingly basic question has remained unresolved until now: when a microRNA precursor is processed into two strands, how does a cell decide which strand to use?
A new study published in Nucleic Acids Research finally provides a clear, data-driven answer. The research shows that microRNA strand selection is not random, as was long assumed. Instead, it follows conserved, programmable rules encoded directly within the structure and sequence of the microRNA itself.
Understanding the MicroRNA Strand Selection Problem
Every microRNA begins life as a precursor molecule that folds into a hairpin structure. This hairpin is then processed into a duplex made up of two strands, commonly referred to as the 5p strand and the 3p strand. Typically, only one of these strands is loaded into the RNA-induced silencing complex (RISC) to guide gene regulation, while the other strand is degraded or plays a minor role.
For years, scientists debated whether this selection was governed by simple thermodynamic chance or by deeper biological rules. While some patterns had been observed, there was no unified framework that could explain strand choice across tissues, developmental stages, and species.
This new study addresses that gap directly.
An AI-Guided Framework Backed by Large-Scale Experiments
The research was led by Marco Mangone, a professor at the Center for Personalized Diagnostics at the Biodesign Institute and the School of Life Sciences at Arizona State University. His team combined large-scale experimental biology with artificial intelligence to decode the logic behind strand selection.
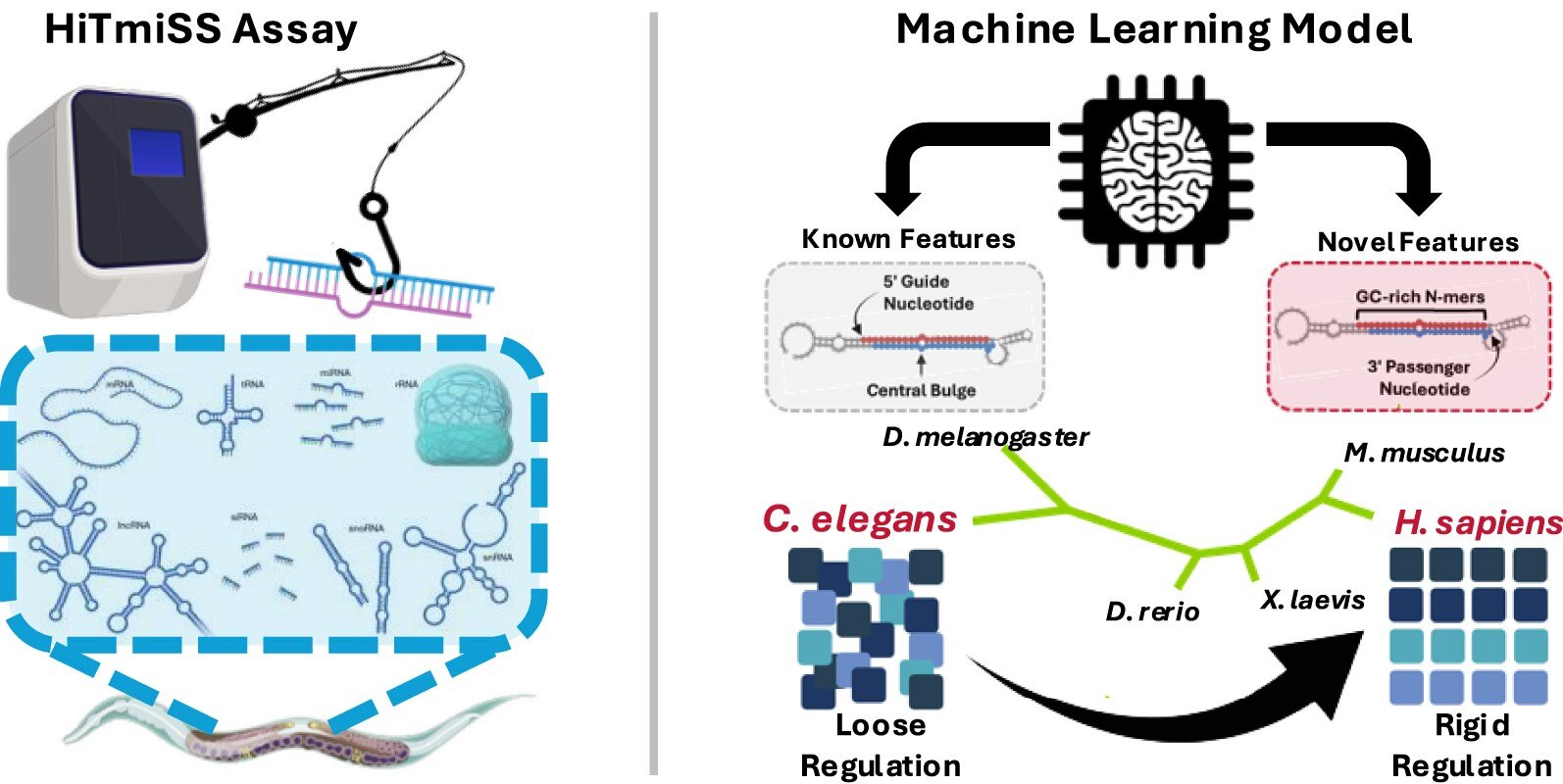
Using the model organism Caenorhabditis elegans, a nematode widely used in genetic research, the researchers developed a new high-throughput experimental method called HiTmiSS. This technique allowed them to precisely measure which microRNA strand was selected across different developmental stages and in specific tissues.
The result was an unprecedented dataset containing thousands of strand-specific measurements, offering a detailed view of how strand usage changes over time and biological context.
Training a Machine Learning Model on 77 Biological Features
To make sense of this massive dataset, the team collaborated with Dr. Heewook Lee, an assistant professor in the School of Computing and Augmented Intelligence at Arizona State University. Together, they trained a machine-learning model designed specifically to predict microRNA strand selection.
What makes this model stand out is its biological grounding. Instead of relying on abstract statistical patterns, it integrates 77 biologically informed features derived from microRNA sequence composition, structural properties, and duplex asymmetry. These features capture subtle aspects of RNA biology that had previously been studied in isolation but never combined into a single predictive framework.
The model proved remarkably accurate. It successfully predicted strand selection not only in C. elegans, but also across a wide range of species, including vertebrates and humans. This cross-species performance demonstrates that the underlying rules of strand selection are deeply conserved through evolution.
Conserved Rules, Tuned by Context
One of the most important conclusions of the study is that strand selection follows conserved rules, but those rules are not rigid. Instead, they can be fine-tuned by developmental stage and tissue context.
In practical terms, this means that the microRNA duplex contains built-in instructions that guide strand selection, but cells can modulate how strongly those instructions are applied depending on where and when the microRNA is expressed. This adds a previously underappreciated layer of control to gene regulation.
The study establishes strand selection as a regulated biological process, rather than a passive byproduct of RNA processing.
Evolutionary Insights Into microRNA Function
The researchers also examined how strand selection has evolved across species. They found a clear distinction between simpler organisms and more complex ones.
In many invertebrates, microRNAs often retain flexible strand usage, meaning both strands may be used under different conditions. In contrast, mammalian microRNAs tend to show a strong bias toward a single dominant strand. This suggests that as organisms evolved greater biological complexity, strand selection became more tightly controlled, likely enabling greater regulatory precision.
These findings help explain how microRNA systems have adapted over evolutionary time to support increasingly complex gene regulatory networks.
Why microRNA Strand Selection Matters
Understanding strand selection is not just an academic exercise. The choice of strand determines which genes are targeted, meaning it can dramatically alter cellular behavior. Misregulation of microRNAs has been linked to cancer, neurological disorders, cardiovascular disease, and many other conditions.
By revealing the rules that govern strand selection, this research provides a foundation for more accurate microRNA-based diagnostics and therapeutic design. If scientists can predict which strand will be active, they can better anticipate downstream gene regulation and avoid unintended effects when designing RNA-based treatments.
Artificial Intelligence as a Tool for Biological Discovery
This study also highlights the growing role of artificial intelligence in biology. Rather than replacing experiments, AI was used here to integrate vast amounts of experimental data into a coherent, predictive framework. The success of this approach demonstrates how machine learning can uncover hidden biological rules that are difficult to detect through traditional analysis alone.
Importantly, the researchers have made all experimental data, prediction scores, and analysis tools openly available, allowing other scientists to apply the model to different species or biological questions.
Expanding the Reader’s Understanding of microRNAs
Beyond strand selection, microRNAs are known to influence nearly every major cellular process, including cell division, development, stress responses, and immune function. Each microRNA can target dozens or even hundreds of genes, making them powerful regulators despite their small size.
The discovery that strand selection itself is regulated adds yet another layer to this already complex system. It reinforces the idea that gene regulation is not governed by single switches, but by multi-layered decision-making processes encoded directly into molecular structures.
A Clear Step Forward in Gene Regulation Research
By combining high-throughput experimentation with AI-driven analysis, this study delivers the first unified, experimentally grounded framework for understanding microRNA strand selection. It answers a long-standing question in molecular biology and opens the door to more precise manipulation of gene regulatory systems.
What was once thought to be a random or secondary detail is now recognized as a fundamental and conserved mechanism of gene control.
Research Paper:
An AI-guided framework reveals conserved features governing microRNA strand selection – Nucleic Acids Research (2026)
https://academic.oup.com/nar/article/54/2/gkaf1510