DNA Shape and Rigidity Play a Decisive Role in How INO80 Controls Gene Expression
Researchers at The University of Texas MD Anderson Cancer Center have uncovered detailed new evidence showing that the physical structure of DNA, particularly its rigidity, directly influences the behavior of the INO80 chromatin remodeling complex. This discovery adds an important new layer to our understanding of how genes are regulated, emphasizing that DNA is not just a sequence of letters but a dynamic material whose shape affects the work of key molecular machines.
The study, published in Molecular Cell and led by Blaine Bartholomew, Ph.D., professor of Epigenetics and Molecular Carcinogenesis, and Shagun Shukla, Ph.D., demonstrates that DNA inflexibility inside the nucleosome can stop INO80 from repositioning chromatin. The findings highlight how DNA bendability—whether it can curve or resist bending—plays a major role in determining where INO80 settles as it organizes and moves nucleosomes.
How INO80 Interacts with DNA
INO80 is one of the major ATP-dependent chromatin remodeling complexes, responsible for sliding nucleosomes along DNA so that genes can be turned on or off as needed. Nucleosomes are the core units of chromatin, formed when DNA wraps tightly around histone proteins. When regions of DNA must be accessed for transcription, replication, or repair, chromatin remodelers like INO80 help open things up.
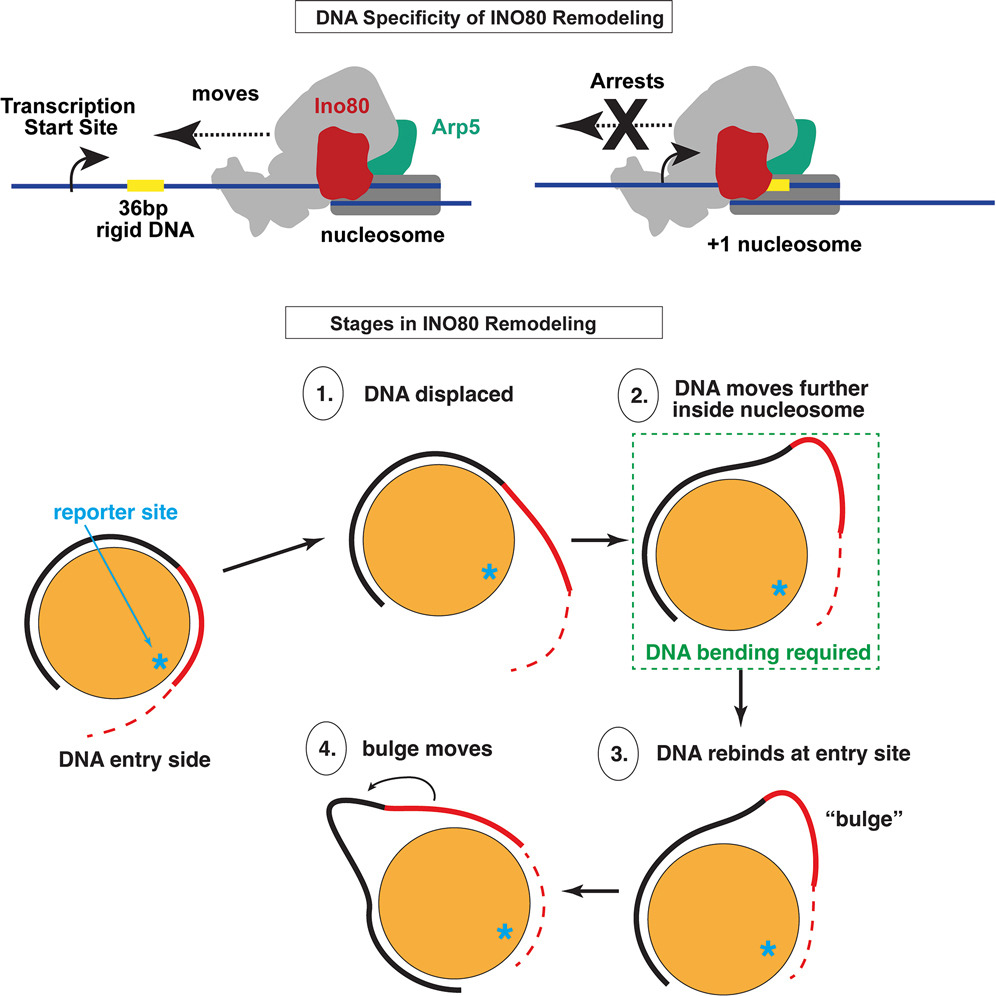
The new study shows that INO80’s behavior is far more selective than previously understood. The team found that when INO80 tries to reposition a nucleosome, it moves DNA not in tiny increments like many other remodelers, but in larger 20–30 base-pair stretches. These movements create temporary loops or bulges in the DNA as INO80 pushes and pulls to slide the nucleosome.
However, INO80 cannot create these bulges in regions where the DNA is especially inflexible. Certain DNA sequences naturally resist bending, and this rigidity prevents INO80 from continuing the remodeling process. As a result, INO80 becomes blocked at these rigid points, which ultimately determines its final position on the nucleosome surface.
This offers a clear explanation for the remarkable specificity of INO80. It positions nucleosomes not just based on genetic information but based on the mechanical properties of DNA, something that earlier research had hinted at without confirming.
DNA Rigidity as a Regulatory Signal
The concept of DNA inflexibility acting as a positioning signal is significant. DNA is often treated as a passive carrier of genetic code, but in reality, it twists, bends, and folds, and these structural characteristics influence how proteins interact with it.
Rigid or inflexible DNA occurs naturally due to certain sequences or biochemical modifications. Previously, scientists suspected that such regions could block the movement of chromatin remodelers like INO80 but lacked detailed evidence. This study presents clear mechanistic insight: rigid DNA disrupts loop formation, halting the wave-like motion INO80 uses to slide the nucleosome.
Because nucleosome positioning is a key factor in controlling which genes are accessible, this rigidity-based blocking mechanism effectively becomes another regulatory layer in gene expression.
The Role of Arp5 as a DNA Shape Sensor
Another important aspect of the study concerns Arp5, one of INO80’s subunits. Earlier research from Bartholomew’s lab showed that Arp5 can rotate dynamically, altering the region where it contacts the nucleosome. In the new study, the researchers observed that Arp5 loses some of its interactions when INO80 is bound to inflexible DNA.
This means the INO80 complex is unable to properly modify or reposition the nucleosome. The researchers propose that Arp5 may act as a kind of sensor, detecting DNA shape and helping regulate whether INO80 proceeds with remodeling or halts at a given location.
This idea of chromatin remodeler subunits acting as structural sensors adds another layer of sophistication to genome regulation.
Why These Findings Matter
The biological importance of this discovery extends beyond basic science. The INO80 complex is involved in DNA transcription, repair, and replication, making it essential for maintaining proper cell function. Disruptions in these processes can lead to severe consequences, including diseases such as cancer.
Indeed, previous studies have shown that mutations, deletions, or overexpression of INO80 and its subunits are associated with both heart disease and cancer. A deeper understanding of INO80’s function offers potential pathways for developing new therapeutic strategies. If INO80’s activity is influenced by DNA shape, then molecules designed to modify DNA rigidity—or mimic its effects—could someday be used to regulate gene expression more precisely.
Gene Expression and DNA Remodeling: Additional Background for Readers
To expand on the context of this study, it helps to explore why chromatin remodeling is such a central theme in molecular biology.
What Is DNA Transcription?
DNA transcription is the first step of gene expression. It involves copying a specific segment of DNA into RNA, which then guides protein production. However, most DNA in cells is tightly packed and inaccessible. Chromatin remodelers like INO80 are essential for temporarily opening up this tightly wound DNA.
Nucleosomes must be repositioned or temporarily ejected for transcription machinery to reach the genetic information it needs. This is why remodelers that control nucleosome placement have such a powerful influence on cellular activity.
What Are Chromatin Remodeling Complexes?
Chromatin remodeling complexes are groups of proteins that use energy from ATP to change how DNA interacts with histones. Different families of remodelers specialize in different tasks:
- Some slide nucleosomes along DNA.
- Some evict nucleosomes entirely.
- Some exchange histone variants.
INO80 belongs to a family known for repositioning nucleosomes with high specificity, particularly during DNA repair and during the regulation of genes that respond to stress.
Why DNA Shape Matters in Modern Genetics
Beyond the letters A, T, C, and G, DNA has physical characteristics—groove width, bendability, stiffness—that influence how proteins recognize and bind it. Recent research across multiple labs has shown that:
- Transcription factors often bind based on shape compatibility as much as sequence.
- DNA rigidity can influence which regions form nucleosomes in the first place.
- Mechanical properties of DNA play a role in replication timing and chromatin domain formation.
The new study adds INO80 to the growing list of molecular systems that rely on DNA mechanics to guide their activity.
A Clearer Picture of Gene Regulation
Together, the findings highlight a sophisticated system in which DNA’s shape, not just its genetic code, influences the organization of chromatin and the regulation of gene expression. INO80’s ability to sense and respond to DNA rigidity strengthens the idea that cells use mechanical cues to coordinate complex genomic processes.
This study not only advances our understanding of chromatin remodeling but also opens the door to new questions about how DNA mechanics contribute to health and disease.
Research Paper:
DNA bendability inside the nucleosome regulates INO80’s nucleosome positioning
https://doi.org/10.1016/j.molcel.2025.10.010