Engineered E. coli Learn to Grow into New Structures Through Direct Cell-to-Cell Contact
A team of researchers at Northeastern University, led by Neel Joshi and Ph.D. student Rong Chang, has developed a fascinating new way to make bacteria grow into complex, living structures—without the need for any external scaffolding. Instead of relying on synthetic supports or natural polymers like cellulose, the team engineered E. coli cells to connect directly with each other through surface proteins, enabling them to form new, controllable shapes.
This achievement, recently published in the Proceedings of the National Academy of Sciences (PNAS, 2025), represents a significant step forward in the field of synthetic biology and bioengineered living materials. It shows that by carefully reprogramming bacteria, we can get them to assemble themselves into structures—similar to how tissues form in nature.
Building Structures from Living Cells
The idea behind this research is surprisingly elegant. Normally, E. coli, a simple and widely studied bacterium, doesn’t naturally form organized, multicellular structures. But Joshi’s team managed to change that by modifying how these cells stick together.
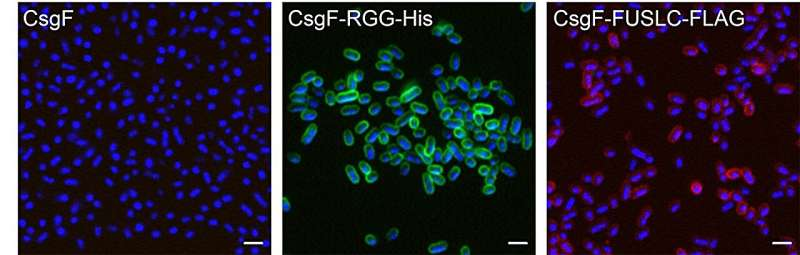
They achieved this by adding an “accessory protein” called CsgF to the outer surface of the bacterial cell. CsgF is typically part of E. coli’s curli system, which helps the bacteria build fibers on their surface. However, in this experiment, CsgF was repurposed as an anchor—a base to which other proteins could attach.
To this anchor, the researchers linked intrinsically disordered proteins (IDPs)—proteins that do not have a fixed 3D shape. These flexible, hair-like proteins were engineered to extend from the cell’s surface. When one engineered E. coli came close to another, these “protein hairs” interacted and bound together, effectively acting like biological Velcro.
By doing this, the cells could form direct physical connections with one another—no external glue or scaffold required. Over time, these interactions caused the bacteria to organize into new multicellular structures.
Reversible and Tunable Cellular Organization
What makes this discovery even more exciting is the reversibility of the system. The team found that by changing environmental factors—such as the buffer composition surrounding the cells—they could control how the bacteria assembled or disassembled.
In simple terms, they could make the cells form a particular shape, break apart, and then re-form that same shape again. This level of control is something that’s rarely been seen before in living systems.
The researchers describe this process as “reversible cellular organization.” It means that the bacteria’s structure isn’t fixed; it’s responsive to the conditions around it. This adaptability could be key to creating dynamic living materials—materials that can change shape, heal themselves, or adapt to their surroundings.
From Scaffolds to Direct Contact
In previous studies, most attempts to create “living materials” involved using scaffolds—frameworks that provide support for growing cells. For example, plants rely on cellulose polymers for rigidity, and even human skin uses keratin as a supportive structure.
However, Joshi’s approach eliminates the need for such scaffolding altogether. The cells themselves are the structure. This direct, cell-to-cell contact approach is much more challenging to achieve but far more flexible.
This is important because in biological systems—like in embryos or growing seeds—cells don’t just multiply randomly. They divide and position themselves in precise ways, forming organized tissues with clear boundaries and orientation. Engineering that kind of controlled growth in bacteria is a complex problem, but Joshi’s lab has taken a meaningful step toward solving it.
The Science Behind the Proteins
The disordered proteins used in this system—such as RGG and FUSLC domains—are key to the process. These proteins don’t have a rigid shape; instead, they move freely and can form weak, reversible bonds with similar proteins on other cells.
When attached to the CsgF anchors, these disordered proteins float around like strands of kelp underwater, ready to interact with proteins on neighboring cells. When they touch, they stick together transiently, forming a mesh-like network that links cells.
Joshi explains that these proteins can be tuned to exhibit different behaviors. For example, by changing which disordered sequence is displayed on the cell surface, researchers can make the bacteria aggregate or separate under certain conditions. This gives scientists a programmable way to control how cells organize—almost like flipping biological switches.
Practical Uses and Future Possibilities
This new platform doesn’t just create interesting biological art—it has potential real-world uses. Joshi and Chang believe that these engineered E. coli could lead to self-growing materials that function like living systems.
For instance, imagine being able to grow materials the way you grow trees for wood. You could let the bacteria self-assemble into strong, organized structures, harvest them, and use them as a sustainable material source.
Beyond that, the researchers think their method could be used to build materials that can survive extreme environments. Some disordered proteins have natural anti-freeze properties, meaning they prevent ice formation. If these proteins are integrated into the bacterial assembly system, the resulting material could remain stable at very low temperatures—or even in high heat.
That opens possibilities for materials that can be used in space exploration, cryogenic storage, or harsh industrial settings.
How This Differs from Traditional Bioengineering
Traditional synthetic biology often focuses on teaching bacteria to make molecules—for instance, using microbes to produce insulin or biofuels. Joshi’s team, however, is interested in teaching bacteria to build structures.
In this work, the researchers aren’t just manipulating gene expression; they’re programming the physical behavior of the cells themselves. That’s a big shift. It takes biology beyond chemistry and into the realm of structural engineering.
Joshi’s lab wants to create living systems that can self-assemble, self-repair, and even change shape on demand. That could revolutionize fields like biomanufacturing, environmental remediation, and tissue engineering.
Challenges and Limitations
While the results are promising, this technology is still in its early stages. The structures formed by these engineered bacteria are relatively simple—mostly aggregates and filament-like assemblies. Creating more complex, three-dimensional architectures remains a major challenge.
There are also questions about mechanical strength, stability, and scalability. Living materials can be fragile, and ensuring they stay viable over time isn’t easy. Moreover, scientists need to ensure these bacteria can’t escape into the environment or interact in unintended ways—a key biosafety concern.
Another issue is specificity. While different disordered proteins can be used to control how cells stick together, ensuring that only the intended interactions occur—and avoiding unwanted clumping—is still difficult.
Even so, Joshi and Chang’s system shows that these challenges are surmountable. Their work provides a foundation for future research on programmable living materials.
The Bigger Picture: Living Materials as the Future
The idea of living materials—substances made of or by living organisms that can grow and adapt—is a rapidly growing field. Scientists imagine building buildings that heal cracks, clothing that repairs itself, or biosensors that respond to toxins in real time.
This E. coli research fits right into that vision. By using disordered proteins and direct cell-to-cell adhesion, it creates a modular and reversible system for building with life.
It’s an early demonstration, but it suggests a future where we won’t just design static materials—we’ll cultivate them.
Research Reference:
Programming Cellular Condensates for Living Materials Using an Intrinsically Disordered Protein Display Platform, Proceedings of the National Academy of Sciences (2025)