Exploring Metabolic Noise Opens New Paths to Better Biomanufacturing
Much like people, microbes don’t always perform at the same level day after day. In the world of industrial biomanufacturing, this inconsistency has been a long-standing frustration. Microbial cells engineered to produce valuable chemicals can be highly productive one moment and surprisingly sluggish the next, even when conditions stay the same. Now, researchers at Washington University in St. Louis have taken a deep dive into this mystery and uncovered what’s really driving these fluctuations — a discovery that could significantly improve how we manufacture bio-based products.
At the heart of this research is a concept known as metabolic noise. This term refers to the differences in metabolic activity seen from one microbial cell to another, even within genetically identical populations. For years, scientists knew this variability existed, but they didn’t fully understand its source or how quickly cells switch between high- and low-productivity states. Without that knowledge, it was difficult to design strategies that consistently favor the most productive cells in large-scale fermentation systems.
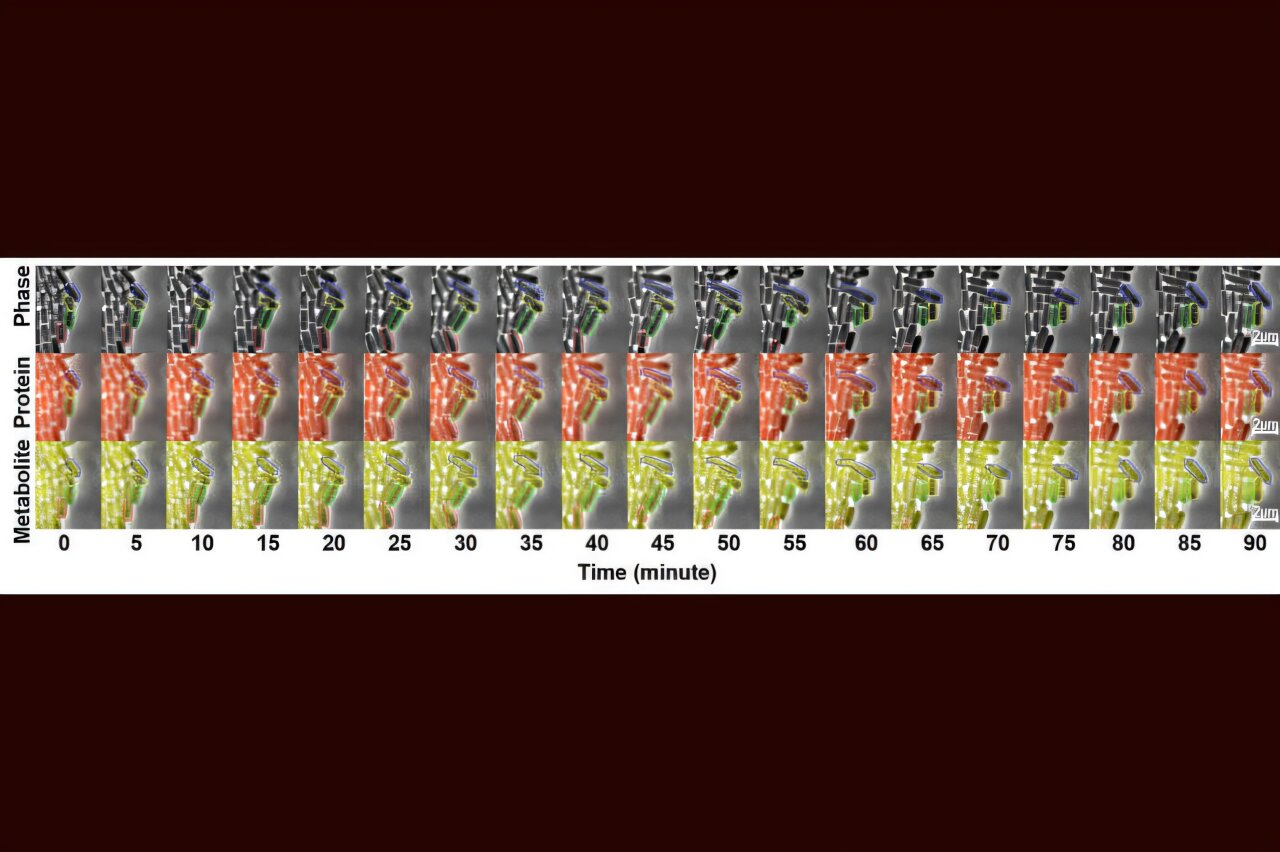
The WashU team tackled this problem by focusing on single-cell behavior, rather than averaging measurements across millions of cells. This shift in perspective turned out to be critical. The study, published in Nature Communications, tracked hundreds of individual Escherichia coli cells as they grew, divided, and carried out normal metabolic functions — all while producing a bright yellow compound called betaxanthin.
Betaxanthin isn’t just visually striking; it played a crucial role in the experiment. The researchers engineered E. coli cells so that betaxanthin production could act as a clear and easily measurable signal of metabolic activity. Unlike many metabolites that are hard to detect inside a living cell, this pigment stands out clearly, making it ideal for studying metabolic fluctuations in real time.
To observe these cells closely, the team built custom microfluidic devices. These tiny platforms allowed them to trap individual bacteria and monitor them continuously over many hours. Using time-lapse microscopy, the researchers could watch how each cell’s production levels changed as it grew and divided, providing a detailed look at metabolic behavior that would be impossible with bulk measurements.
What they found was striking. Betaxanthin production wasn’t stable at all. Individual cells frequently switched between high-production and low-production states, sometimes within just a few hours. This rapid switching explained why even well-designed microbial strains can behave unpredictably during fermentation.
Digging deeper, the researchers identified the main driver behind this variability. Roughly 50% of the observed metabolic noise came from fluctuations in the number of enzyme molecules responsible for making betaxanthin. These enzyme fluctuations weren’t caused by changes in the environment or genetic mutations, but by the inherent randomness of gene expression. In other words, even under identical conditions, cells naturally produce different amounts of the same enzyme at different times.
Interestingly, cell growth_setting played a much smaller role than many scientists expected. Variations in growth rate accounted for less than 10% of the total betaxanthin variability. This finding challenges the common assumption that faster-growing cells are always more productive and highlights enzyme expression as a far more important factor.
Armed with this new understanding, the team went a step further. They didn’t just want to explain metabolic noise — they wanted to use it to their advantage. Using experimental data from the single-cell measurements, the researchers developed computational models to test different strategies for increasing overall production. They evaluated four possible control approaches, asking which ones could push a microbial population toward higher output.
The models pointed to a promising solution: enrich cells that naturally overproduce the key enzyme. Because enzyme expression fluctuates randomly, some cells temporarily make more enzyme than others. If those cells could be given a growth advantage, they would gradually dominate the population and boost total production.
To test this idea, the researchers engineered a gene circuit that links higher enzyme expression to faster growth. Cells that stochastically produce more of the betaxanthin-making enzyme gain a competitive edge, grow more quickly, and produce even more of the desired compound. When the team tested this approach in fermentation experiments, the results matched the model’s predictions, leading to a substantial increase in betaxanthin output.
Beyond betaxanthin, the implications of this work are broad. Many industrial bioprocesses rely on microbes to produce pharmaceutical ingredients, dietary supplements, biodegradable plastics, and biofuels. In all of these systems, metabolic noise can limit efficiency and increase costs. By identifying enzyme copy number fluctuations as a dominant source of variability, this research provides a clear target for improving microbial performance.
This work also fits into a larger push toward sustainable, zero-waste manufacturing. Biomanufacturing is often seen as a greener alternative to traditional chemical production, but inefficiencies can still lead to wasted resources. Keeping microbial “workers” focused and productive is essential for making these processes economically and environmentally viable.
Understanding Metabolic Noise in Simple Terms
Metabolic noise might sound abstract, but it’s a fundamental property of life at the molecular level. Inside every cell, genes are constantly being turned on and off, and molecules are made and degraded in probabilistic ways. This randomness means that no two cells are ever doing exactly the same thing at the same time, even if they’re genetically identical and living side by side.
In many cases, cells can tolerate or even benefit from this variability. It allows populations to adapt quickly to changing environments. However, in industrial settings where consistency and high output are key, metabolic noise becomes a challenge. The WashU study shows that understanding this noise — rather than trying to eliminate it entirely — can lead to smarter engineering strategies.
Why Single-Cell Studies Matter
Traditional biomanufacturing research often looks at average behavior across millions or billions of cells. While useful, this approach hides the extreme differences that exist between individual cells. Single-cell techniques, like the microfluidic methods used in this study, reveal a much richer picture of microbial life.
As tools for single-cell analysis continue to improve, researchers expect to uncover more hidden sources of variability and new ways to control them. This could lead to more robust microbial strains and more predictable industrial processes.
In the end, this research highlights an important shift in thinking. Instead of treating metabolic noise as an obstacle, scientists can now see it as an opportunity — one that opens new paths toward more efficient and reliable biomanufacturing.
Research paper:
https://www.nature.com/articles/s41467-025-67733-1