Open-Source Robotic System Cuts Manual Cell Culture Time by 61% While Boosting Seeding Consistency
Researchers have developed an open-source robotic system that significantly reduces the manual workload involved in cell culture while also improving how evenly cells are seeded across microplates. Published in PNAS Nexus, the study presents a practical and relatively affordable automation solution aimed at one of the most routine yet labor-intensive tasks in biomedical research: cell passaging and seeding.
At its core, the system demonstrates that automation does not have to be prohibitively expensive or locked behind proprietary software. Instead, it shows how thoughtfully designed open-source tools can deliver measurable improvements in efficiency, reproducibility, and researcher well-being.
What the system does and why it matters
Cell culture is foundational to modern life-science research. Whether scientists are studying gene expression, drug responses, or cellular behavior under a microscope, they rely heavily on maintaining healthy and consistently seeded cell populations. However, traditional cell culture workflows are time-consuming, physically demanding, and often variable, especially when performed manually with pipettes.
The newly developed system, called the Automated Cell Culture Splitter (ACCS), directly addresses these issues. It automates the process of passaging cells—moving cells from one plate to another once they become too dense—while precisely controlling how many cells are seeded into each well of a 96-well microplate.
According to the researchers, the system reduced hands-on time by 61% compared to manual passaging. While the overall processing time was longer due to robotic operation, the amount of time that required direct human involvement was dramatically lower. For busy labs handling multiple experiments, this distinction is critical.
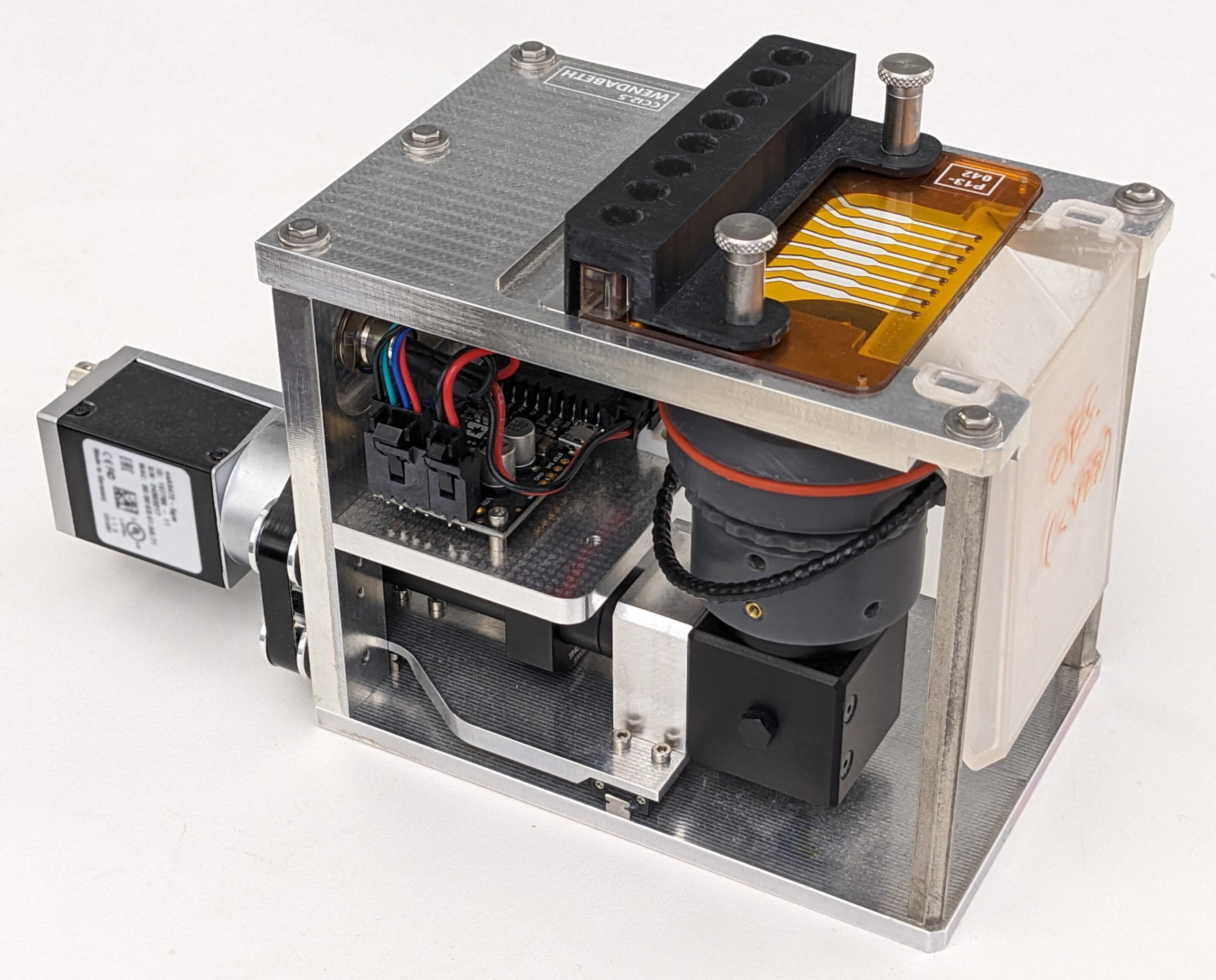
The technology behind the automation
The Automated Cell Culture Splitter is built around the widely used Opentrons OT-2 liquid handling robot, a benchtop automation platform already familiar to many research labs. What makes this system unique is its integration with a custom-built Cell Counting Imager (CCI), designed specifically to measure cell concentrations accurately before seeding.
The Cell Counting Imager is essentially an inverted darkfield microscope paired with a custom flow cell. It can measure eight samples simultaneously, allowing the system to calculate exactly how much cell suspension is needed to seed a specified number of cells into each destination well.
Key technical features include:
- A custom flow cell with eight chambers, mounted on top of the imaging system
- A motorized linear stage that moves the optical system to image each chamber
- A row of ports aligned so that the OT-2’s multichannel pipette can inject samples from one column of a source plate at a time
By integrating imaging directly into the workflow, the system avoids reliance on rough visual estimates—one of the biggest sources of variability in manual cell culture.
Tested on a widely used cell line
To evaluate performance, the researchers tested the system using HEK293T cells, a human embryonic kidney cell line commonly used in molecular and cell biology. This choice makes the results especially relevant, as HEK293T cells are a staple in many laboratories worldwide.
In direct comparisons between automated and manual workflows, the differences were clear. Plates seeded by the automated system showed higher consistency in cell density across wells than those seeded manually. Manual seeding relied on visual estimation and hand pipetting, which naturally introduces variation from well to well.
The benefits became even more apparent in high-throughput imaging experiments. Plates prepared using the automated system achieved a 92% rate of usable imaging sites, compared to just 52% for the best manually seeded plate. For experiments where image quality and uniformity are critical, this improvement alone could justify adopting automation.
Cost and accessibility
One of the most striking aspects of the system is its relatively low cost. The authors estimate that the full setup can be built for approximately $18,000, which is substantially cheaper than many commercial cell culture automation platforms that can cost several times more.
Equally important, the system is fully open-source. The hardware designs, software, and documentation are publicly available, allowing labs to adapt the system to their own needs. This openness lowers the barrier to entry and encourages collaboration, customization, and further innovation.
For smaller academic labs or research groups with limited funding, this approach offers a realistic path toward automation without sacrificing transparency or control.
Impact on researcher health and productivity
Beyond efficiency and data quality, the authors also point to an often-overlooked benefit: researcher health. Manual pipetting, especially during repetitive tasks like cell passaging, is associated with work-related musculoskeletal disorders, including wrist, shoulder, and neck strain.
By reducing the need for continuous manual handling, the automated system could help minimize these risks while also freeing researchers to focus on experimental design, data analysis, and interpretation rather than repetitive bench work.
Why seeding consistency matters
Uniform cell seeding is more than a technical detail—it directly affects experimental outcomes. Variations in cell density can influence cell behavior, growth rates, gene expression, and response to treatments. Inconsistent seeding can introduce noise that masks real biological effects, making experiments harder to reproduce.
Automation helps standardize this step. By combining precise cell counting with controlled liquid handling, the system ensures that each well starts with nearly the same number of cells. This level of consistency is particularly important for high-content imaging, drug screening, and other assays that rely on comparing results across dozens or hundreds of wells.
The broader trend toward open-source lab automation
This work fits into a growing movement toward open-source laboratory automation. As hardware costs fall and software tools become more accessible, researchers are increasingly building custom systems tailored to their workflows rather than relying solely on commercial solutions.
Open-source automation offers several advantages:
- Transparency, allowing researchers to understand and verify how systems work
- Flexibility, enabling customization for specific experimental needs
- Community-driven improvement, where users share updates, fixes, and enhancements
The Automated Cell Culture Splitter is a clear example of how this approach can deliver real-world benefits without compromising scientific rigor.
Looking ahead
While the system currently focuses on 96-well microplates and routine passaging tasks, its modular design suggests it could be expanded to support other formats or workflows in the future. With continued development and community involvement, similar systems could become standard tools in research labs aiming to balance efficiency, cost, and reproducibility.
In a field where small inconsistencies can lead to large experimental differences, this work highlights how thoughtful automation can improve both the science and the daily experience of doing research.
Research paper:
Open-source cell culture automation system with integrated cell counting for passaging microplate cultures – PNAS Nexus
https://academic.oup.com/pnasnexus/article/4/12/pgaf385/8405882