Researchers Expand the Human Genome Map to 2.37 Million Regulatory DNA Elements
Scientists have taken a major step toward understanding how the human genome actually works by dramatically expanding the known map of regulatory DNA. A research team led by Zhiping Weng, Ph.D., and Jill Moore, Ph.D. at UMass Chan Medical School has nearly tripled the number of identified regulatory DNA elements, pushing the total to 2.37 million. This new resource represents the largest and most detailed map yet of the DNA sequences that control when, where, and how genes are switched on or off in human cells.
The findings were published in the journal Nature in January 2026 and are part of the long-running Encyclopedia of DNA Elements (ENCODE) project, a global scientific effort that has been mapping functional elements of the genome for more than two decades.
What Are Regulatory DNA Elements and Why Do They Matter?
When people think about DNA, they often focus on genes that code for proteins. However, only a small fraction of the human genome actually encodes proteins. The vast majority of our DNA plays a regulatory role, helping control how genes behave rather than acting as instructions themselves.
These regulatory sequences are known as cis-regulatory elements (CREs). They are typically located outside protein-coding regions and are responsible for controlling gene transcription. In simple terms, they act as genetic switches, determining whether a gene is turned on, turned off, or adjusted up or down in activity.
CREs are essential for:
- Cell specialization
- Developmental processes
- Responses to environmental signals
- Maintaining normal biological function
Many diseases are now known to be linked not to broken genes, but to misregulated genes, making these regulatory elements critically important for understanding human health.
A Massive Expansion of the Regulatory Map
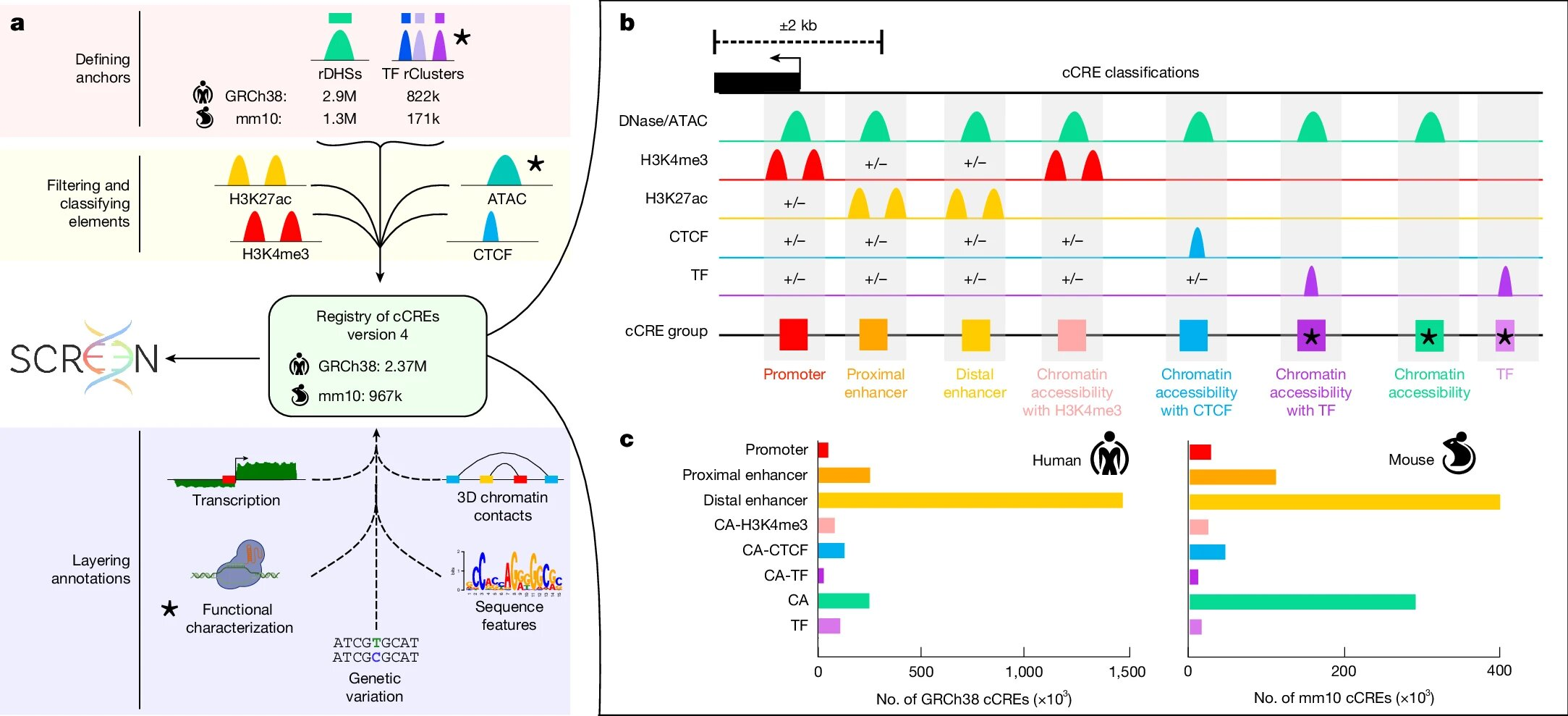
Before this study, scientists had identified roughly 900,000 regulatory elements in the human genome and about 300,000 in the mouse genome. The new research expands these numbers to:
- 2.37 million candidate cis-regulatory elements (cCREs) in humans
- 927,000 cCREs in mice
This expansion was not just about counting more DNA regions. It involved systematic functional annotation, meaning the researchers gathered evidence that these sequences actually show biochemical signs of regulatory activity.
According to the research team, this new registry is the most comprehensive catalogue of regulatory DNA ever assembled, offering unprecedented insight into the noncoding regions of the genome.
How Scientists Built the Largest Regulatory Registry Ever
One of the most important aspects of this work is how the registry was created. Instead of relying on a single method, the researchers integrated data from a wide range of high-throughput biological assays.
In total, the study analyzed:
- 5,712 human experiments
- 758 mouse experiments
All of these experiments were generated by the ENCODE Consortium and include data such as:
- Chromatin accessibility measurements
- Histone modification patterns
- Transcription factor binding profiles
- Other biochemical signals associated with gene regulation
By combining these datasets with improved computational analysis, the researchers were able to functionally characterize over 90% of known human regulatory elements. This represents a major leap in coverage compared to earlier genome annotations.
Enhancers, Silencers, and Dual-Function DNA
Regulatory elements are often categorized based on their function. The expanded registry captures a broad range of regulatory behaviors, including:
- Enhancers, which increase gene expression
- Silencers, which repress gene expression
- Promoter-associated elements
- Elements linked to specific transcription factor binding
One of the most interesting findings from this study is that some regulatory elements do not have a fixed role. The same DNA sequence can act as an enhancer in one cell type and a silencer in another. This context-dependent behavior depends on which transcription factors are present in a given cell.
This discovery highlights the flexibility and complexity of gene regulation and explains why predicting gene behavior based on DNA sequence alone has been so challenging.
Cell Type Specificity and Biological Context
Another key advance is the ability to see where and when regulatory elements are active. Not all cCREs operate in every cell. Many are active only in specific tissues, developmental stages, or disease-related states.
The updated registry provides a reference atlas that shows:
- Which regulatory elements are active
- In which cell and tissue types
- Under which biological conditions
This level of detail is essential for understanding complex biological systems and how gene regulation changes during development or disease progression.
Connecting Regulatory DNA to Human Disease
One of the most powerful uses of this expanded map is in interpreting genome-wide association studies (GWAS). GWAS have identified tens of thousands of genetic variants linked to common diseases such as:
- Heart disease
- Diabetes
- Schizophrenia
The challenge is that most of these variants lie outside protein-coding genes, making it difficult to understand how they influence disease risk.
By overlaying GWAS signals onto the cCRE registry, researchers can now:
- Identify which regulatory elements contain disease-associated variants
- Determine which genes those elements likely control
- Focus on the relevant cell types for each condition
A Concrete Example: Red Blood Cell Development
The study includes a detailed example involving red blood cell traits. Using the expanded regulatory map, researchers analyzed genetic variants associated with red blood cell characteristics.
Rather than assuming the nearest gene was responsible, the team examined which genes were functionally regulated by the relevant cCREs. This approach pointed to KLF1, a gene that acts as a central regulator of red blood cell development.
Experimental disruption of one regulatory region reduced KLF1 activity, supporting the conclusion that genetic variation in this region affects red blood cell traits primarily through KLF1. This example demonstrates how regulatory maps can move researchers beyond guesswork to mechanistic understanding.
The Role of the ENCODE Project
This work is part of the broader ENCODE project, a two-decade international effort organized into four successive five-year phases. ENCODE aims to systematically identify all functional elements in the human and mouse genomes.
Zhiping Weng has served as a principal investigator in all four ENCODE phases and has led the consortium’s data analysis center for the past decade, helping coordinate data integration and public release.
The new cCRE registry reflects years of collaborative effort and sets a new benchmark for genomic resources.
Why This Matters for the Future of Genomics
By combining dense regulatory maps, functional assays, and human genetics, this expanded registry provides a foundational tool for:
- Studying gene regulation
- Understanding developmental and cell-specific programs
- Interpreting noncoding genetic variation
- Clarifying how genetic differences contribute to disease
The registry is publicly accessible through an updated online portal, making it a valuable resource for researchers worldwide.
As scientists continue to explore the noncoding genome, this work brings us closer to understanding how the vast majority of our DNA shapes who we are, how our cells function, and why diseases develop.
Research paper:
https://www.nature.com/articles/10.1038/s41586-025-09909-9