Scientists Map the Oat Pangenome Unlocking Genetic Secrets for Better, Healthier Crops
Oats have long been a staple for those who care about nutrition and heart health, but despite their popularity, oat breeding has lagged behind other major cereal crops like wheat, rice, and maize. The reason? Their genome is extraordinarily large and complex, making genetic improvement a slow process. Now, a groundbreaking study published in Nature in 2025 is changing that. An international team of researchers, led by scientists from Brigham Young University (BYU) and other global institutes, has unveiled a comprehensive oat pangenome and pantranscriptome—a monumental step forward in understanding this fascinating grain.
What the New Research Achieved
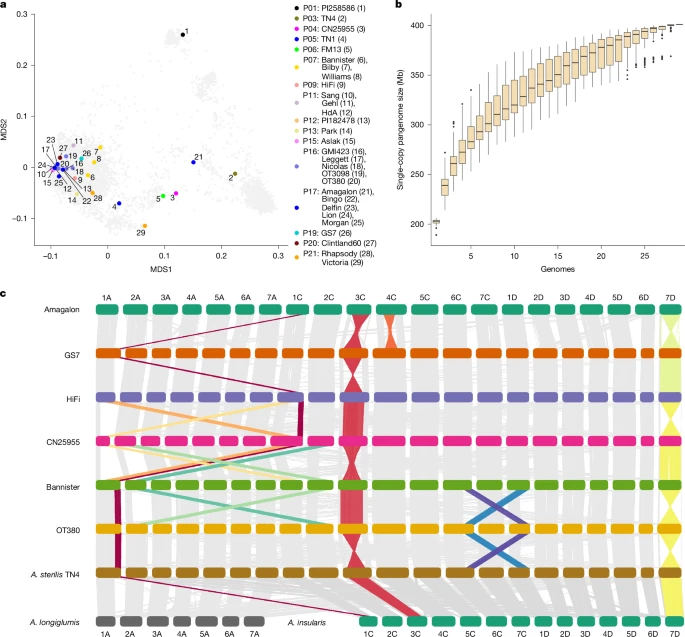
The study, titled “A pangenome and pantranscriptome of hexaploid oat,” presents the first large-scale genomic reference that captures the full spectrum of oat diversity. Researchers analyzed 33 oat lines, including both wild and domesticated varieties, creating a pangenome—a complete catalog of all genes present across different oat types.
While the first oat genome was sequenced back in 2022, this new pangenome reveals how that genetic story changes from one variety to another. It distinguishes core genes (shared across all oats) from accessory genes (unique to certain types), offering a much richer picture of oat biology.
To go even further, the team developed what they call a pantranscriptome—an atlas showing which genes are active in different parts of the oat plant, such as leaves, roots, and seeds, and at various developmental stages. Using BYU’s DNA Sequencing Center, the researchers studied gene expression across six tissue types and stages in 23 oat lines, creating one of the most detailed maps of oat gene activity ever compiled.
The Hidden Complexity of the Oat Genome
Oats are hexaploid, meaning they have six sets of chromosomes originating from three ancestral genomes. This level of complexity makes their genome much larger and more intricate than most cereals. The new research shows that this genomic complexity contributes to why breeding oats has been more challenging.
The study revealed that the oat genome is highly dynamic—chromosomes frequently undergo structural rearrangements such as inversions (segments flipping in orientation) and translocations (segments moving to new locations). These rearrangements can affect how traits are inherited and how easily breeders can cross different varieties.
The researchers also identified that gene loss in one subgenome is often compensated for by upregulated genes in others. This genetic balancing act helps oats remain stable and adaptable despite their tangled chromosomal history.
Why Structural Rearrangements Matter
The researchers discovered that these structural changes in the oat genome are not just random quirks—they have real implications for crop improvement. For example, some chromosomal inversions were linked to early flowering traits, while other rearrangements influenced plant height and drought tolerance.
Such findings are crucial because they explain why oat breeding progress has historically been slow. When chromosomes rearrange, recombination (the process that mixes genes during breeding) can be disrupted, making it harder to combine desirable traits.
Understanding these rearrangements allows breeders to plan crosses more strategically, avoiding genetic combinations that are unlikely to produce stable, fertile offspring.
A Second Study Expands the Global Picture
Alongside the Nature paper, a companion study published in Nature Communications by the same international team analyzed around 9,000 oat samples collected from all over the world. This research looked at the population structure of both wild and cultivated oats, identifying how genetic diversity is distributed across regions and how specific chromosome structures are linked to environmental adaptation.
The findings revealed that wild oats (Avena sterilis) consist of at least four distinct genetic populations, while the cultivated oats (Avena sativa and Avena byzantina) also form separate clusters. Some of the same chromosomal rearrangements observed in the pangenome study were found to play roles in local adaptation, suggesting that these structural variations help oats survive and thrive in specific climates.
This global-scale analysis confirmed that oat evolution involved multiple domestications and complex polyploid origins, meaning the modern oat plant is the product of repeated hybridization events and gene exchanges across species.
Why This Research Is a Game-Changer
With these new resources—the pangenome, pantranscriptome, and global diversity dataset—breeders now have powerful tools to accelerate oat improvement. These data sets allow scientists to identify genes tied to important traits like nutritional quality, disease resistance, drought tolerance, and yield.
For instance, traits linked to beta-glucan, a soluble fiber known for reducing cholesterol and supporting heart health, can now be prioritized in breeding programs. Similarly, genes influencing flowering time or root development can be targeted to create varieties better suited for different environments.
The research also helps explain why oats have historically underperformed in yield improvement compared to crops like wheat. Structural genome barriers limited the success of traditional breeding. Now, by understanding these barriers at the DNA level, breeders can make data-driven decisions to overcome them.
The Power of Collaboration and Technology
This breakthrough is a result of global scientific collaboration involving experts in gene sequencing, plant genetics, and bioinformatics from multiple countries. BYU professors Rick Jellen and Jeff Maughan, both leaders in plant and wildlife sciences, played pivotal roles.
The project used high-throughput DNA sequencing and advanced bioinformatics tools to assemble chromosome-scale genomes—an impressive technical feat given the size and complexity of oat DNA. The BYU DNA Sequencing Center provided the infrastructure for large-scale gene activity studies, which were essential for building the pantranscriptome.
Researchers emphasized that such massive projects are only possible through international cooperation, with contributions from universities, crop research institutes, and genome centers worldwide.
Why Oats Deserve More Attention
Beyond this research, it’s worth appreciating why oats are such an important crop in the first place. They’re naturally rich in beta-glucan, protein, fiber, and essential minerals like magnesium and zinc. Unlike wheat and barley, oats contain avenanthramides, unique antioxidants known for their anti-inflammatory properties.
Oats are also more tolerant to poor soils and cool climates, making them a promising crop for sustainable agriculture in regions facing climate stress. They are increasingly used as a base for plant-based milk, snack bars, and breakfast cereals, contributing to a growing global market focused on health and sustainability.
However, until now, the lack of deep genomic understanding has limited the crop’s improvement. With the pangenome and pantranscriptome available, researchers can finally leverage modern genomic tools—like genome-wide association studies (GWAS) and genomic selection—to breed better oat varieties faster.
What This Means for Future Crop Breeding
The new data sets offer not just a blueprint for oats but a model for how complex polyploid crops can be studied and improved. Many other cereals and oilseeds have similarly complex genomes, and the methods used here—especially combining genomic and transcriptomic data—could be applied to those crops as well.
For countries like India, where oats are gaining traction as both a human and animal feed crop, this research provides a valuable foundation for breeding locally adapted varieties. By using the genetic information from global oat collections, Indian breeders can identify germplasm with the right traits for heat tolerance, drought resilience, and nutritional enrichment.
The Road Ahead
These studies answer long-standing questions about oat evolution, domestication, and genetic barriers. More importantly, they open new pathways for improving a crop that fits perfectly into modern demands for healthier, sustainable foods.
The research highlights how understanding diversity at the genetic level can drive agricultural progress. Oats, once seen as a secondary crop, might soon take center stage in efforts to produce food that’s good for both people and the planet.
Research Reference:
Avni, R. et al. (2025). A pangenome and pantranscriptome of hexaploid oat. Nature. DOI: 10.1038/s41586-025-09676-7