Sulfolobus islandicus Research Expands the Genetic Toolkit for Biotechnology and Drug Delivery Applications
Sulfolobus islandicus may not be a household name, but in the world of biotechnology and synthetic biology, this tiny archaeal organism is quietly becoming a big deal. A new study from researchers at the University of Illinois Urbana-Champaign sheds light on how this extreme microbe can be genetically engineered more precisely than ever before. The work focuses on discovering and validating chromosomal integration sites in Sulfolobus islandicus, a breakthrough that significantly expands its usefulness as a platform for industrial biotechnology, metabolic engineering, and even future drug delivery systems.
The findings were published in the journal Trends in Biotechnology and represent a detailed effort to overcome one of the biggest limitations in working with this organism: the lack of well-defined, reliable locations in its genome where new genes can be safely and stably inserted.
Why Sulfolobus islandicus Matters in Biotechnology
Sulfolobus islandicus is an archaeon, a domain of life distinct from bacteria and eukaryotes. What makes it especially interesting is its ability to thrive in extreme environments, particularly at very high temperatures (around 70–85°C) and highly acidic conditions (pH 2–4). These traits make it naturally resistant to contamination and well suited for industrial processes that would destroy most conventional microbial hosts.
Because of these characteristics, S. islandicus has long been viewed as a promising candidate for synthetic biology, where microbes are redesigned to produce useful chemicals, enzymes, or materials. However, progress has been slow compared to more common organisms like E. coli or yeast, largely due to limited genetic tools.
The Core Problem: Lack of Defined Integration Sites
While several genetic tools already exist for Sulfolobus islandicus, researchers have struggled with a major bottleneck: the absence of well-characterized chromosomal integration sites. Without these, inserting genes into the genome can be unpredictable, potentially disrupting essential functions or leading to unstable gene expression.
Stable chromosomal integration is critical for long-term industrial applications. Plasmid-based expression systems, which are common in bacteria, are often unreliable under extreme growth conditions. This makes chromosomal integration the preferred option—but only if suitable sites are known.
Using CRISPR-COPIES and Multi-Omics to Map the Genome
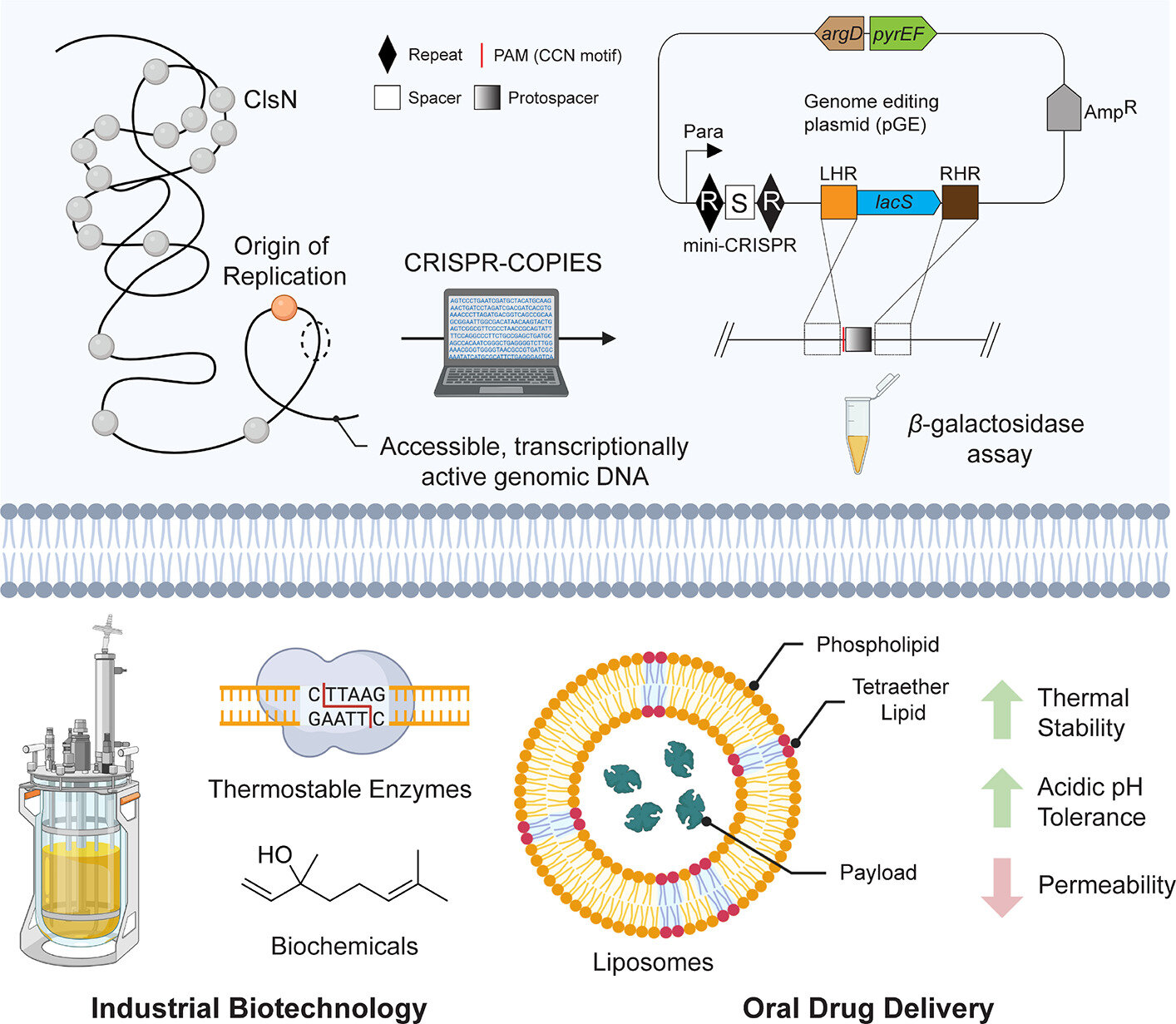
To solve this problem, the research team employed a sophisticated pipeline known as CRISPR-COPIES, combined with a multi-omics strategy that integrated both genomic and epigenomic data. This approach allowed the researchers to scan the S. islandicus genome and identify regions that could tolerate gene insertion without harming the organism.
Their analysis focused on intergenic regions, stretches of DNA located between genes, which are less likely to interfere with essential cellular processes. Using this strategy, the team identified 66 CRISPR RNAs (crRNAs) that target 21 distinct intergenic regions with strong potential as chromosomal integration sites.
In Vivo Validation Using the Native CRISPR System
Identifying candidate sites computationally was only the first step. The researchers then moved on to experimental validation, using the organism’s own Type I-A CRISPR-Cas system to perform precise genome editing.
To test whether these sites actually worked in living cells, they used a lacS reporter system, which measures β-galactosidase activity. This allowed them to quantify how well genes inserted at different sites were expressed.
Out of the 66 predicted crRNAs, 13 crRNAs corresponding to eight chromosomal sites were successfully validated in vivo. These sites supported stable gene integration and measurable gene expression, confirming their suitability for genetic engineering.
An important finding here was the presence of positional effects. In simple terms, the location of a gene in the genome significantly influenced how strongly it was expressed. This insight is especially valuable for future metabolic engineering efforts, where precise control over gene expression levels is often critical.
A Practical Demonstration: Engineering Membrane Lipids
To demonstrate the real-world potential of their newly identified integration sites, the researchers carried out a proof-of-concept experiment. They overexpressed a gene called GrsB, short for glycerol dibiphytanyl glycerol tetraether ring synthase B, at one of the validated chromosomal sites.
GrsB plays a key role in the biosynthesis of GDGT lipids, which are unique ether-linked membrane lipids found in many archaea. These lipids are known for their exceptional stability under extreme conditions.
The result of GrsB overexpression was a clear change in membrane composition, specifically an increase in the number of cyclopentane rings within the lipid molecules. This modification can significantly affect membrane rigidity, permeability, and thermal stability.
Why Archaeal Lipids Are So Interesting
Archaeal lipids, including GDGTs, differ fundamentally from bacterial and eukaryotic membrane lipids. They are ether-linked rather than ester-linked and often form monolayer membranes, which are far more stable at high temperatures and low pH.
Because of these properties, archaeal lipids are attracting interest for drug delivery applications, such as the design of highly stable liposomes capable of surviving harsh environments. By gaining genetic control over lipid biosynthesis in Sulfolobus islandicus, researchers open the door to custom-engineered membranes with tailored properties.
Expanding the Genetic Toolbox for Non-Conventional Hosts
One of the broader impacts of this work is its contribution to the growing effort to move beyond traditional microbial hosts. While organisms like E. coli and yeast are well understood, they are not always ideal for industrial processes.
This study significantly strengthens Sulfolobus islandicus as a robust, non-conventional host by providing:
- Well-defined chromosomal integration sites
- A validated framework for predictable gene expression
- Compatibility with endogenous CRISPR-Cas systems
- Tools that support long-term genetic stability
Together, these advances make S. islandicus a more practical option for industrial biotechnology, metabolic engineering, and synthetic biology research.
Data, Reproducibility, and Community Impact
The researchers have made their work highly accessible to the scientific community. Supporting datasets, including information on integration sites, CRISPR targets, and experimental validation, have been deposited in public repositories. This transparency allows other labs to quickly adopt and build upon the new tools.
By combining computational prediction with rigorous experimental validation, the study sets a strong example for how genetic toolkits can be expanded in other archaeal and non-model organisms.
Looking Ahead
This research marks an important step toward turning Sulfolobus islandicus into a reliable cellular factory. With defined integration sites now available, future studies can focus on more ambitious goals, such as multi-gene pathway engineering, large-scale metabolite production, and the creation of novel biomaterials.
As interest grows in resilient biological systems capable of operating under extreme conditions, Sulfolobus islandicus is likely to play an increasingly visible role.
Research paper:
https://doi.org/10.1016/j.tibtech.2025.11.003