Thirty New Fungal Lineages Discovered Through eDNA Long-Read Sequencing
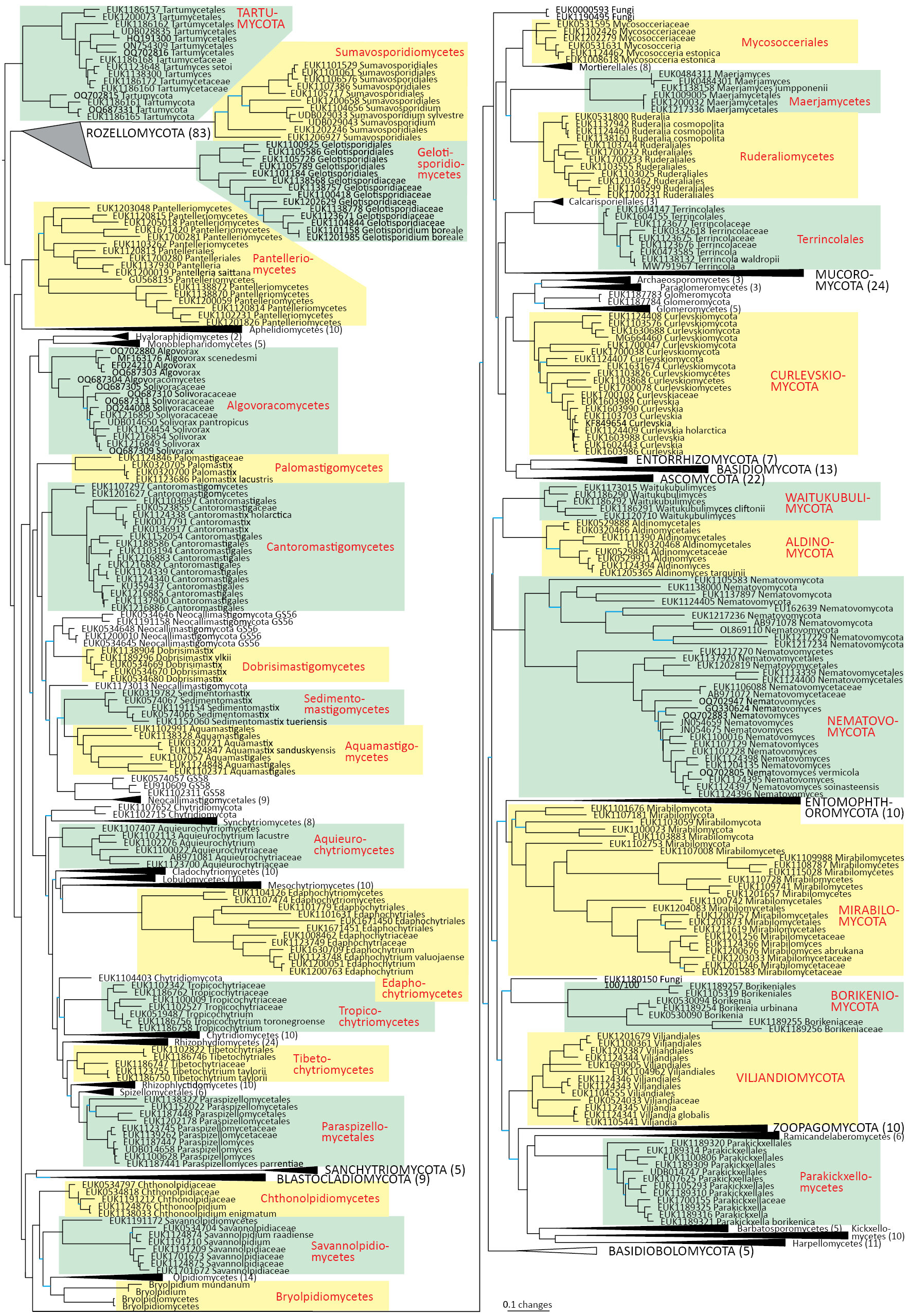
A team of international researchers led by Prof. Leho Tedersoo from the University of Tartu, Estonia, has uncovered a remarkable find — 30 previously unknown fungal lineages, some at the order, class, and even phylum levels. The study, recently published in the journal MycoKeys, represents one of the biggest leaps in fungal taxonomy in recent years, using nothing more than environmental DNA (eDNA) and cutting-edge long-read sequencing technology.
What the Researchers Did
The team analyzed a large dataset known as the EUKARYOME long-read database, which contains high-quality DNA sequences from eukaryotic organisms found in environmental samples such as soil, water, and sediments. These sequences are often referred to as “dark taxa” — genetic fingerprints of organisms that are known to exist from their DNA but have never been isolated, cultured, or visually observed.
To make sense of these elusive organisms, the researchers focused on the ribosomal RNA (rRNA) operon, specifically the SSU–5.8S–LSU regions, which are highly useful for reconstructing evolutionary relationships among eukaryotes. Using long-read sequencing allowed them to capture long, continuous DNA fragments that preserve more information for accurate classification — something that short-read methods often fail to do.
By combining phylogenetic analyses with a fresh approach to taxonomy, the researchers systematically organized these sequences into 30 entirely new fungal lineages. These discoveries span from new orders to new phyla, expanding the fungal kingdom’s known diversity by nearly one-third.
How They Classified the New Fungi
The key innovation of the study was not just in finding new DNA sequences, but in how these sequences were formally classified. Because many of these organisms have never been seen or cultured, traditional taxonomy — which depends on physical specimens or holotypes — couldn’t be applied.
To address this, Tedersoo and colleagues proposed two new taxonomic concepts:
- Nucleotype – a DNA sample derived from a holotype specimen.
- Legitype – a DNA sequence that can act as a type specimen itself when a physical specimen doesn’t exist or is lost.
This marks an important shift in taxonomy, opening the door to naming and describing organisms that exist only in genetic form — a necessary step for cataloging life that cannot be grown or observed in a lab.
Some of the Newly Described Lineages
Among the newly identified fungal lineages, several have already been given formal names. A few examples include:
- Pantelleria saittana (in the new class Pantelleriomycetes)
- Paraspizellomyces parrentiae (in the new order Paraspizellomycetales)
- Aquieurochytrium lacustre (in the new class Aquieurochytriomycetes)
- Edaphochytrium valuojaense (in the new class Edaphochytriomycetes)
- Tibetochytrium taylorii (in the new class Tibetochytriomycetes)
- Tropicochytrium toronegroense (in the new class Tropicochytriomycetes)
- Aquamastix sanduskyensis (in the new class Aquamastigomycetes)
In total, these lineages represent a wide range of evolutionary branches, suggesting that the fungal kingdom is far more diverse than previously recognized.
How the Team Named the Taxa
An interesting detail of the study is that the names of the new taxa were chosen collaboratively by all co-authors. The team decided to honor the type localities where these DNA sequences originated, incorporating native language stems — including Amerindian, Sámi, and Estonian linguistic roots — into the names.
This naming approach reflects both scientific precision and cultural respect, acknowledging the diverse regions and communities associated with the samples.
The Taxonomic Approach and Phylogeny
To verify the placement of these new lineages within the fungal kingdom, the team constructed maximum-likelihood phylogenetic trees using the SSU–5.8S–LSU regions. These trees revealed where each new lineage sits in relation to existing fungal classes and phyla.
Many of the new taxa were found to branch deeply within the fungal tree, occupying positions that are distinct from all previously known groups. This suggests that entire lineages of fungi — representing hundreds of millions of years of evolution — have gone unnoticed until now simply because they could not be cultured or seen.
The authors also developed diagnostic nucleotide signatures to distinguish certain groups at the genetic level. For example, the family Edaphochytriaceae was defined based on a unique nucleotide pattern found in its LSU region.
Why This Discovery Matters
This work is much more than a taxonomy exercise. It fills major gaps in the fungal tree of life and gives researchers formal names and classifications for groups that were previously invisible.
Environmental sequencing has shown for years that the majority of microbial diversity remains undescribed. Now, with long-read sequencing and new taxonomic standards, scientists can finally start naming and organizing these mysterious organisms.
By doing so, they can:
- Better understand ecosystem functions, since fungi play key roles in nutrient cycling, symbiosis, and decomposition.
- Improve biodiversity assessments, as many ecological studies depend on accurate species-level data.
- Provide a foundation for future biotechnological discoveries, since new fungal lineages may hold novel enzymes or metabolites.
This kind of work demonstrates that even at high taxonomic levels — like phylum and class — there are still huge swaths of biodiversity left to uncover.
A Glimpse into the World of Environmental DNA
Environmental DNA (eDNA) refers to genetic material that organisms shed into their surroundings — from spores, skin cells, waste, or decomposing bodies. By extracting and sequencing this DNA from soil, water, or air, scientists can detect species without ever seeing or capturing them.
For microorganisms, which are often too small or rare to culture, eDNA provides a powerful tool for understanding biodiversity. The challenge, however, is linking sequences to actual organisms. Many eukaryotic sequences have remained unclassified because they don’t match anything in existing databases.
That’s where long-read sequencing comes in. It generates longer, more complete DNA fragments, giving researchers a better chance to reconstruct evolutionary relationships and confidently place new organisms on the tree of life.
What We Still Don’t Know
While the discovery is groundbreaking, there are still many unknowns. These fungi exist only as DNA data points — no one has observed their morphology, physiology, or ecology. Their roles in nature remain a mystery. Are they decomposers, symbionts, or parasites? Do they interact with plants or animals?
There’s also ongoing debate about using DNA sequences as type material in taxonomy. Some scientists argue that without physical specimens, the full picture of an organism is incomplete. Others believe this is the only realistic way to describe much of life on Earth, especially the microbial majority.
Despite the controversy, the study sets a precedent. It shows that naming life from sequences alone can be done responsibly, with rigorous analysis and transparent standards.
Broader Implications
The work by Tedersoo and his team opens new doors for microbial taxonomy. It also hints at how much more there is to discover. If 30 entirely new fungal lineages can be identified from a single large database, imagine what remains hidden in other ecosystems — from tropical soils to deep-sea sediments.
The study serves as a reminder that our understanding of life’s diversity is still incomplete. Fungi, in particular, are crucial to ecosystems but remain among the least studied groups of organisms. With tools like eDNA and long-read sequencing, we are finally starting to see the outlines of this hidden world.
Research Reference:
Leho Tedersoo et al., “Thirty novel fungal lineages: formal description based on environmental samples and DNA,” MycoKeys (2025). DOI: 10.3897/mycokeys.124.161674