Quantum Mechanical Molecular Fingerprints Are Transforming How Machine Learning Understands Molecules
Machine learning has become a powerful tool in chemistry, but one long-standing problem has been figuring out the best way to “describe” a molecule so a computer can truly understand it. A new study from chemists at Cornell University offers a major breakthrough by showing that molecules don’t need to be described only by their atomic structure. Instead, using their quantum mechanical fingerprints—specifically the behavior of electrons—can dramatically improve how accurately machine learning models predict molecular properties.
The research introduces a new method called Semi-Local Density Fingerprints (SLDFs), which captures essential quantum information about a molecule in a form that machine learning models can efficiently use. According to the researchers, this approach can deliver predictions that are up to 100 times more accurate than those produced using commonly used techniques.
Why Describing Molecules Is a Big Deal in Machine Learning
In chemistry-focused machine learning, the way a molecule is represented—often called a molecular descriptor—is critical. Most existing models rely on structural data: which atoms are present, where they are located, how far apart they are, and what angles they form. For example, a water molecule is typically described as one oxygen atom bonded to two hydrogen atoms at a specific angle and bond length.
This approach works reasonably well, but it has clear limitations. Structural descriptions can miss subtle energetic and electronic effects that play a huge role in how molecules behave. As a result, even sophisticated machine learning models may struggle to predict properties accurately, especially for molecules they have never seen before.
The Cornell team, led by Robert DiStasio, wondered why machine learning models should rely only on simplified structural information when chemists routinely calculate detailed quantum mechanical data. That question led directly to the development of SLDFs.
What Are Semi-Local Density Fingerprints?
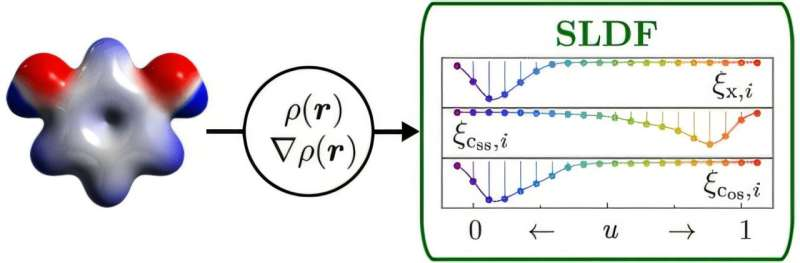
Semi-Local Density Fingerprints are molecular descriptors built from electron density, a fundamental quantum mechanical quantity. Electron density describes the probability of finding electrons at different locations around a molecule. Since electrons govern bonding, reactivity, and energy, this information captures what truly matters at the most basic level.
To create SLDFs, the researchers first perform a Density Functional Theory (DFT) calculation. DFT is one of the most widely used quantum mechanical methods in chemistry and materials science, balancing accuracy with computational efficiency. These calculations typically take only a few minutes on modern computers.
Once the DFT calculation is complete, the electron density data is processed into a compact, fixed-length fingerprint that can be fed directly into a machine learning model. Importantly, these fingerprints are designed to be invariant to rotations, translations, and permutations of atoms, which makes them ideal for machine learning applications.
Testing SLDFs on Molecular Energies
In this study, the researchers focused on predicting molecular conformational energies. Conformational energy refers to the energy differences between various shapes or arrangements of the same molecule. For instance, even a simple molecule like water can exist in slightly distorted states, each with a different energy.
The team trained machine learning models using datasets containing thousands of molecular conformations. The real challenge, however, was not predicting energies for molecules already included in the training data. Instead, the researchers tested whether the models could accurately predict conformational energies for entirely new molecules.
The results were striking. Models using SLDFs consistently outperformed both traditional machine learning descriptors and standard DFT calculations. In many cases, the SLDF-based models achieved accuracy improvements of orders of magnitude, demonstrating that quantum-informed descriptors provide a far richer and more transferable representation of molecular behavior.
Solving the Transferability Problem in Machine Learning
One of the biggest mysteries in molecular machine learning is transferability—the ability of a model trained on one set of molecules to make accurate predictions for very different molecules.
In this study, the researchers trained their machine learning models only on molecules composed of first- and second-row elements from the periodic table, such as hydrogen, carbon, nitrogen, and oxygen. Remarkably, the same models were able to accurately predict conformational energies for molecules containing third-row elements like sulfur and phosphorus.
This level of transferability is difficult to achieve with structure-based descriptors alone. SLDFs succeed because they focus on electrons, which are present in all molecules regardless of which elements are involved. By learning from electron density rather than atomic labels, the machine learning model captures universal physical principles instead of surface-level patterns.
Why Electron Density Makes Such a Difference
Electron density is at the heart of quantum chemistry. It determines how atoms bond, how molecules interact, and how energy flows through chemical systems. By using electron density as a molecular fingerprint, SLDFs allow machine learning models to learn directly from quantum mechanical reality rather than approximations based on geometry alone.
Another key advantage is efficiency. While high-level quantum calculations can be extremely expensive, SLDFs rely on standard DFT calculations, which are already widely used in research and industry. This makes the approach practical for large-scale applications.
Broader Implications for Chemistry and Materials Science
The potential applications of SLDFs extend far beyond conformational energy prediction. Because the method is applicable to any system containing electrons, it can be used across chemistry, physics, and materials science.
Current and future applications include:
- Predicting reaction energies
- Estimating reaction barriers, which relate directly to reaction speed
- Modeling molecular properties such as stability and electronic behavior
- Assisting in the design of new molecules and materials with specific target properties
By combining quantum mechanical insight with machine learning efficiency, SLDFs offer a powerful new tool for accelerating discovery in fields ranging from catalysis to drug development.
How This Work Fits into the Bigger Picture of AI in Science
This research highlights an important lesson for scientific machine learning: better data representations can be just as important as better algorithms. Rather than relying solely on more complex neural networks, the Cornell team showed that embedding physical meaning directly into the input features can dramatically improve performance.
SLDFs provide a clear example of how physics-informed machine learning can outperform purely data-driven approaches. As machine learning continues to expand across scientific disciplines, methods like this are likely to play a key role in making models more accurate, reliable, and interpretable.
The Research Behind the Breakthrough
The study was led by Zhuofan Shen, a doctoral student in chemistry and chemical biology, with Robert A. DiStasio Jr. serving as the corresponding author. Additional contributors included Zachary M. Sparrow and other members of the DiStasio research group at Cornell University.
The findings were published in The Journal of Physical Chemistry Letters in December 2025, marking a significant step forward in the integration of quantum chemistry and machine learning.
Research Paper:
Learning Molecular Conformational Energies Using Semi-Local Density Fingerprints
https://doi.org/10.1021/acs.jpclett.5c02222