A New Low-Cost T-Cell Sequencing Technique Lets Scientists Analyze Millions of Immune Cells at Once
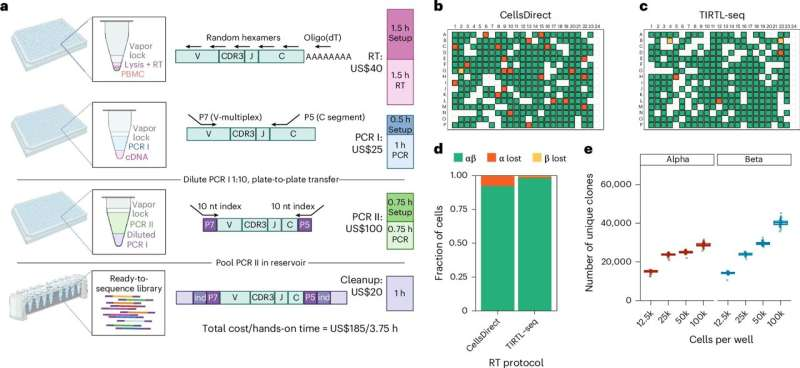
A major breakthrough from St. Jude Children’s Research Hospital is reshaping how researchers study T cells—the immune cells responsible for recognizing infections and tumors. Scientists at St. Jude have introduced a new technique called TIRTL-seq (Throughput-Intensive Rapid TCR Library sequencing), which can analyze an entire T-cell repertoire with unmatched depth, accuracy, and affordability. This method doesn’t just slightly improve existing technology—it dramatically expands what’s possible in immune-system research.
T cells rely on specialized proteins called T-cell receptors (TCRs) to detect threats in the body. Each TCR has two complementary parts, and understanding which two halves pair together is crucial for decoding how the immune system recognizes disease. The challenge is that a single person can have millions of distinct T cells, each with unique TCR combinations. Historically, analyzing these cells has required expensive, low-throughput methods that limit researchers to studying only thousands of cells at a time. That’s a tiny fraction of the true diversity present in the immune system.
St. Jude’s new method directly tackles these limitations. According to the team, TIRTL-seq provides the most complete view of a person’s T-cell repertoire ever achieved—at only about 10% of the cost of conventional approaches. The technique was published in Nature Methods and is positioned to make T-cell research far more accessible to labs around the world.
Researchers demonstrated that TIRTL-seq can accurately process up to 30 million T cells in a single run, whereas the most common existing methods top out at around 20,000 cells. The cost difference is equally striking: traditional sequencing typically runs about $2,000 for 20,000 cells, but TIRTL-seq costs around $200 for 10 million cells. In other words, scientists can now examine vastly more data for a fraction of the price.
The method mixes automation, more efficient laboratory techniques, and streamlined computational analysis to bring costs down without sacrificing accuracy. One key innovation is that scientists split samples into many subsamples. This allows statistical pairing of TCR chains that would otherwise require expensive single-cell sequencing platforms. By combining these subsamples intelligently, researchers can reconstruct which TCR halves belong together across millions of cells.
The St. Jude team initially built TIRTL-seq to reduce the cost and workload of large-scale TCR studies launched in 2023—specifically those aimed at understanding catastrophic childhood diseases. The method surpasses benchmarks set by all competing technologies. Not only is it cheaper and deeper, but it is also quantitatively more accurate, especially when identifying rare TCRs that may reveal early immune responses or signs of hidden infections.
To demonstrate the method’s potential, scientists applied TIRTL-seq to a real-world clinical dataset: blood samples from the St. Jude Tracking Study of Immune Responses Associated with COVID-19 (SJTRC). This study began in 2020 during the pandemic, with the goal of understanding how people’s immune systems react to SARS-CoV-2 over time.
Using TIRTL-seq, researchers successfully tracked changes in T-cell receptors before and after a participant contracted COVID-19. The new method revealed immune-system signatures that traditional sequencing would have missed. Its analysis was so comprehensive that it flagged signs of a previously undetected Epstein–Barr virus (EBV) infection in the individual’s immune history. The fact that TIRTL-seq could identify a separate, non-COVID infection based solely on T-cell patterns shows just how powerful this approach may become for future diagnostics.
St. Jude has made the entire technique freely available online, complete with step-by-step instructions, so that any lab with basic equipment can adopt the method. This move aligns with the researchers’ goal of “democratizing immune memory research” by removing price barriers.
The release of TIRTL-seq matters beyond this single study. It represents a shift toward large-scale, highly affordable T-cell repertoire sequencing, something that has long been considered the most limiting factor in immunology. A complete map of TCR diversity gives scientists a deeper understanding of how immune memory forms, how infections shape immune responses, and how diseases like cancer distort immune patterns. With millions of T cells analyzed at once, researchers can study immune biology at a level that simply wasn’t feasible before.
Because T cells play roles in infections, cancer, autoimmunity, vaccine responses, and more, the applications are broad. The ability to obtain accurate TCR pairing information also supports the growing field of T-cell-based therapies, which require identifying receptors that respond to specific targets.
Below is additional background that may help readers appreciate why this technology is such a leap forward.
Understanding T Cells and T-Cell Receptors
T cells act as the immune system’s surveillance force. Each T cell carries a unique T-cell receptor, which is responsible for recognizing fragments of pathogens or abnormal cells. A TCR consists of two chains—commonly called the alpha chain and the beta chain. The exact pairing of these chains determines what each T cell recognizes.
Scientists often want to know:
- how many different TCR combinations exist in a person
- how these combinations change after infection or vaccination
- whether specific TCRs respond to cancer cells
- whether unusual TCR patterns signal an autoimmune condition
Traditional sequencing techniques can reveal the diversity of these chains but usually fail to correctly pair them, meaning researchers cannot tell which alpha and beta belonged to the same T cell. Some cutting-edge single-cell platforms can obtain this pairing but are too expensive for large studies.
This is where TIRTL-seq stands out: it provides true chain pairing at massive scale and low cost.
Why Scale Matters in T-Cell Research
The human body contains tens of millions of unique T cells at any given time. Studying only a few thousand is like examining just one page of a massive encyclopedia.
Large-scale TCR sequencing enables:
- detection of rare T cells responding to early infection
- tracking immune changes during disease progression
- identifying immune signatures that predict how a patient will respond to therapy
- mapping immune memory over a lifetime
TIRTL-seq’s ability to process 30 million cells at once helps shift immune research from small snapshots to complete landscapes.
A Step Toward Immune-Based Diagnostics
One of the most exciting implications of this research is that the immune system itself records a person’s history of infections. TIRTL-seq’s detection of an unnoticed EBV infection hints at a future where clinicians might diagnose diseases by reading T-cell patterns—a kind of immune fingerprint.
Such diagnostics could:
- catch infections earlier
- reveal chronic or latent viral activity
- support personalized treatment decisions
While the technology isn’t yet approved as a diagnostic tool, the new findings show that T-cell repertoire analysis has genuine clinical potential.
Open Access and Scientific Progress
St. Jude’s decision to make TIRTL-seq openly available is another major step forward. When methods remain locked behind proprietary platforms, only well-funded labs can explore the immune system in detail. By contrast, open protocols allow:
- smaller labs to conduct high-impact immunology studies
- large consortia to generate standardized datasets
- rapid expansion of T-cell research across different diseases
Open access accelerates discovery—and in fields like immunology, faster discovery can translate into new treatments and better outcomes for patients.
Research Paper:
TIRTL-seq: deep, quantitative and affordable paired TCR repertoire sequencing
https://doi.org/10.1038/s41592-025-02907-9