A Single Prime Editing System Shows Promise for Treating Many Different Genetic Diseases
A new gene-editing strategy developed at the Broad Institute is generating excitement because it aims to tackle a major challenge in modern genetic medicine: the need to design a separate therapy for nearly every individual mutation. The approach, called PERT (Prime Editing-mediated Readthrough of premature termination codons), was created by a team led by gene-editing pioneer David Liu. What makes PERT stand out is its ambition to address a wide range of diseases caused by nonsense mutations, all with a single editing system.
Nonsense mutations interrupt protein production early by inserting a premature termination codon—essentially a rogue stop signal. These mutations can occur in many different genes, and they collectively account for about 24% of the 200,000 pathogenic mutations in the ClinVar database. Because they appear across thousands of diseases and patients, finding a universal way to bypass them has long been a major goal.
Rather than correcting each individual mutation—an expensive, time-intensive process—PERT takes a broader route. It installs a specialized tool inside cells that lets them read through faulty stop signals and continue making full-length, functional proteins. This could one day lead to a one-time treatment useful for many unrelated genetic disorders, especially rare ones that currently receive little attention because therapies are too costly to develop one by one.
How PERT Works and Why It’s Different
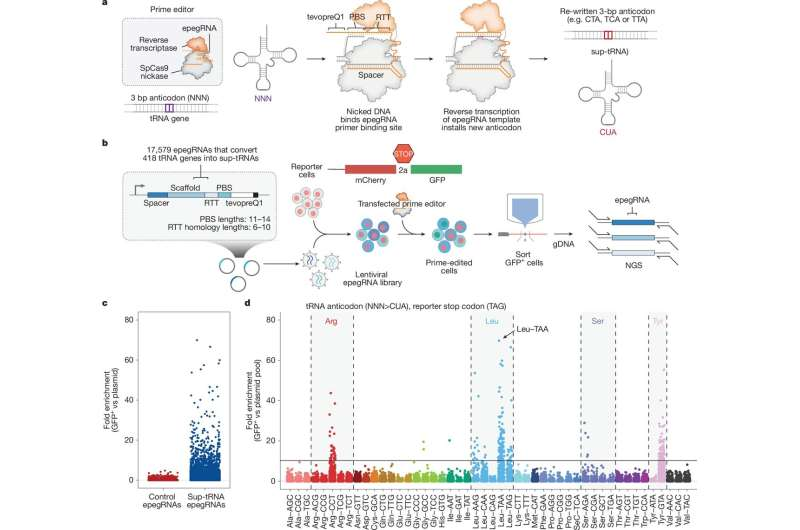
The core of PERT builds on prime editing, a precise DNA-editing method introduced by Liu’s lab in 2019. Traditional prime editing can make targeted DNA changes without requiring double-stranded breaks, but each application still requires designing a new edit for each mutation. PERT instead uses prime editing to install an engineered suppressor tRNA, or sup-tRNA, directly into the genome.
Here’s why that matters. When a cell builds a protein, it reads messenger RNA (mRNA) three bases at a time. Normally, when it reaches one of three specific codons (UAA, UAG, or UGA), the cell stops building the protein. If a nonsense mutation introduces one of these codons too early, the result is a truncated protein that often cannot function. A suppressor tRNA, however, can “override” that early stop by inserting an amino acid instead, letting the protein production continue.
The Broad team didn’t simply insert an extra tRNA into the cell. Instead, they replaced one of the body’s redundant tRNA genes with a newly engineered version. This ensures the edited tRNA is produced at natural levels, reducing risks seen in older approaches where overexpressing suppressor tRNAs disrupted normal protein synthesis.
This required enormous experimentation: the team tested tens of thousands of tRNA variants before finding a highly efficient suppressor. Once optimized, the prime editing system was able to install this sup-tRNA reliably across cells.
Lab Results in Multiple Disease Models
The researchers tested PERT in human cell models of Batten disease, Tay–Sachs disease, and Niemann–Pick disease type C1—each caused by different nonsense mutations in different genes. In all cases, the new editor restored enzyme activity to 20% to 70% of normal levels. These levels are believed to be high enough to reduce or even avoid symptoms in many genetic disorders.
The team also tested PERT in a mouse model of Hurler syndrome, a severe lysosomal storage disorder. After editing, tissues in the brain, liver, and spleen showed about 6% of normal enzyme activity, which was still strong enough to dramatically reduce visible disease signs in the mice.
A key concern with any genome-editing technology is unintended consequences, such as off-target edits or interference with healthy protein production. According to the study, none were detected. The engineered suppressor tRNA also appeared at low, natural levels, which may help explain why it didn’t disrupt normal stop codons or cause unwanted protein extensions.
The researchers highlighted that mammalian cells have several backup mechanisms ensuring proper protein synthesis, which may further reduce potential risks.
Why This Could Transform Genetic Medicine
Developing gene-editing therapies today often requires spending years and millions of dollars per disease, which limits commercial feasibility. Companies must choose which conditions to pursue, leaving countless others without any treatment prospects. PERT represents an attempt to break that cycle by focusing on a shared biological problem found across many disorders.
If one editing system could treat large groups of patients, it could drastically increase the number of diseases with viable therapies. It would also give companies and researchers better incentives to invest in treatments that currently affect only small populations.
The team behind PERT is now working on improving the technology and testing it across additional animal models. Their long-term goal is to bring PERT into clinical trials, and to inspire more research into broad-applicability, disease-agnostic gene-editing tools.
Additional Background: Why Nonsense Mutations Are Such a Big Deal
Among all types of genetic mutations, nonsense mutations are especially disruptive. Unlike missense mutations (which swap one amino acid for another) or synonymous mutations (which change DNA without altering the protein), nonsense mutations terminate protein synthesis prematurely. This often leaves cells with nonfunctional molecules, leading directly to disease.
Some well-known conditions involving nonsense mutations include certain cases of:
- Cystic fibrosis
- Duchenne muscular dystrophy
- Hemophilia
- Metabolic and lysosomal storage disorders
Treating nonsense mutations is challenging because every gene—and every mutation—requires its own tailored solution. Over the past two decades, researchers have tested readthrough drugs, antisense therapies, and customized gene editing, but none have offered a universal option. PERT is the first strategy aiming to restore protein function across many diseases without needing mutation-specific edits.
Additional Background: What Makes Prime Editing Unique
Prime editing is often described as a “search-and-replace” system for DNA. Unlike CRISPR–Cas9, which cuts both DNA strands, prime editing uses a nickase enzyme paired with a specialized guide RNA that contains instructions for the desired edit. This allows precise changes without relying on error-prone repair pathways.
Since its debut, prime editing has been considered one of the most versatile tools in genome engineering. However, its promise has been limited by the same hurdle facing every editing technology: the need to tailor each treatment to each mutation. PERT shows how prime editing might be used more broadly, not just as a surgical tool but as a way to install general-purpose molecular upgrades in cells.
Looking Ahead
While the findings are promising, PERT is still in early development. Questions remain about long-term safety, whether more types of suppressor tRNAs will be needed, and how efficiently the editor can be delivered throughout the human body. Delivery is often the biggest obstacle in genetic medicine—especially for organs like the brain.
Still, the early data suggests that even partial restoration of enzyme activity can significantly improve disease outcomes. If further testing confirms safety, PERT may become one of the most important steps toward truly scalable genetic therapies.
Research Paper:
Prime editing-installed suppressor tRNAs for disease-agnostic genome editing
https://www.nature.com/articles/s41586-025-09732-2