Genetic Sequences Found to Direct Epigenetic Changes in Plants, Revealing How DNA Methylation Really Begins
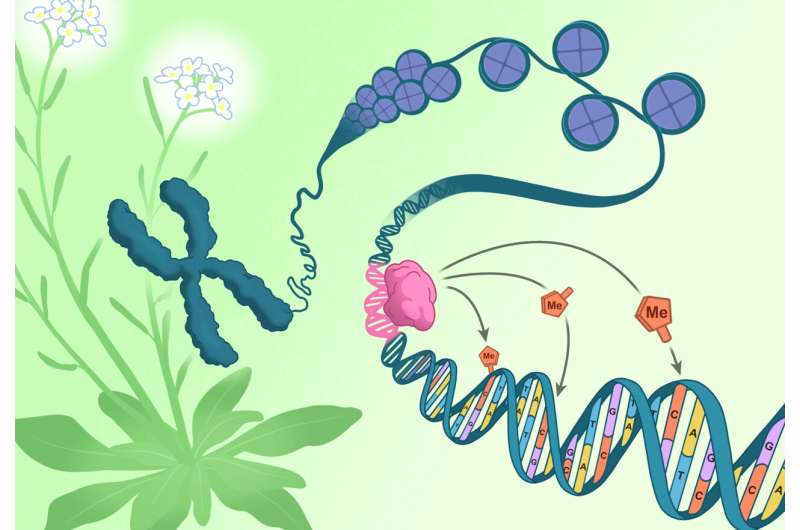
Scientists at the Salk Institute have uncovered a major shift in our understanding of how epigenetic patterns form, answering a long-standing mystery: if epigenetics regulates genes, what regulates epigenetics itself? Their new research shows that specific DNA sequences—not just existing epigenetic marks—can instruct where DNA methylation, a key epigenetic modification, is placed. This discovery reshapes how we think about gene regulation, development, and the potential for future epigenetic engineering.
Epigenetics refers to chemical tags that sit on DNA or on the proteins that package DNA. These tags help determine which genes are active or silent, and they shape the identity of cells in plants, animals, and humans. One of the most important epigenetic tags is DNA methylation, where a methyl group is added to particular cytosine bases in DNA. When a region is methylated, it is usually silenced, preventing gene expression or blocking the activity of wandering genetic elements called transposons, which can otherwise damage genome stability.
Even though every cell holds the same genetic code, epigenetic patterns differ dramatically from one cell type to another. These differences make it possible for specialized tissues—like roots, leaves, neurons, or liver cells—to exist. Errors in these methylation patterns can cause developmental issues in both plants and animals, and in mammals they are linked to diseases such as cancer. Because of this, understanding where methylation comes from and how it is targeted is a major goal in biology.
Until now, researchers mostly knew how methylation is maintained—that is, how existing methylation marks are re-copied after cell division to preserve the original pattern. What they did not understand was how new methylation patterns arise in the first place. Without a mechanism to create new patterns, it would be impossible for organisms to develop, respond to stress, or regenerate tissue properly.
The new Salk Institute study fills this knowledge gap using Arabidopsis thaliana, a small flowering plant commonly used in research. Arabidopsis handles disruptions in its epigenetic machinery much better than animals do, making it ideal for experiments that would be too risky in mammalian systems. In this plant, a group of proteins known as CLASSYs (four in total) help guide methylation machinery to different genomic sites. One of these proteins, CLASSY3, has long been known to act in reproductive tissues, but scientists did not understand why it targets some locations and not others.
To solve this, the Salk team used a forward genetic approach to search for factors that influence CLASSY3 activity. They discovered a set of transcription factors that they named RIMs. These RIM proteins belong to a large broader family called REPRODUCTIVE MERISTEM (REM) transcription factors. What made the discovery remarkable is that these RIM proteins bind specific DNA sequences, and those sequences are essential for directing methylation to nearby genomic regions. When researchers altered or removed the DNA sequences where RIMs normally dock, the methylation pathway failed completely. For the first time, scientists had clear proof that genetic information itself can guide epigenetic modification.
The mechanism works like this: RIMs attach to their corresponding DNA sequences. Once they are docked, they help recruit CLASSY3 and methylation-related machinery, including components of the RNA-directed DNA methylation (RdDM) pathway. This pathway generates small RNAs that guide methyltransferases to tag specific cytosines with methyl groups. Because different reproductive tissues express different combinations of RIM proteins, each tissue generates a unique methylation pattern. These patterns would not exist without the DNA-binding instructions provided by RIM transcription factors.
The research team went further by showing that changing where a RIM protein is expressed can redirect the entire methylation machinery to new genomic locations. This demonstrates not just correlation, but direct control, revealing that DNA sequences can act like addresses telling the cell where to install methylation marks.
The same issue of Nature Cell Biology also featured a complementary study from researchers at UCLA, who used reverse genetics to identify additional REM family members involved in DNA methylation targeting. Together, both studies strongly support the idea that genetic and epigenetic layers are more interconnected than previously believed.
As senior researcher Julie Law explains, earlier models assumed methylation only begins where previous epigenetic modifications already existed. That theory could not explain how new epigenetic states emerge during development or environmental stress. Now we know that DNA itself provides instructions, offering a second, previously hidden regulatory layer. This insight has far-reaching implications. For example, if DNA sequences can recruit methylation machinery, scientists could theoretically design DNA motifs that intentionally target or repair epigenetic patterns. This could lead to new approaches in crop engineering, improving resilience or productivity, or even potential medical strategies for conditions influenced by improper methylation.
Understanding epigenetic targeting is especially important because methylation helps silence transposons—genetic elements capable of jumping around the genome. Transposon activity can weaken plant fitness and cause genomic instability in animals, including humans. The ability to engineer precise methylation could offer new ways to keep such elements under control.
This discovery also opens many new scientific questions. Researchers still do not know how widespread DNA-sequence-driven methylation is in other plant tissues or whether something similar might occur in animals. Because the REM family is large, it is likely that more RIM-like proteins exist, and identifying them could reveal a much broader system controlling epigenetic patterns. Scientists also wonder how these genetic signals interact with other influences, such as chromatin structure, environmental changes, or developmental cues.
To help readers better understand the broader context, it’s useful to include a quick overview of foundational concepts related to the study.
What Is DNA Methylation?
DNA methylation is a biochemical process where a methyl group is attached to a cytosine base in DNA. In both plants and animals, methylation often leads to gene silencing. In plants, this mechanism is essential not only for normal development but also for preventing transposons from becoming active. Plants have a more varied and abundant set of methylation pathways than most animals, which is one reason they are excellent models for studying epigenetic regulation.
What Are Transposons and Why Do They Matter?
Transposons, sometimes called “jumping genes,” are mobile DNA elements that can copy or move themselves within the genome. While sometimes beneficial in evolution, they often disrupt genes or destabilize genomes. Methylation acts like a shield, keeping these elements silent. When methylation fails, transposons can become active, leading to stress responses or developmental abnormalities.
Why Arabidopsis thaliana Is a Powerful Research Model
Arabidopsis has become the go-to plant for molecular biology research because:
- Its genome is small and fully sequenced.
- It has short generation times.
- It tolerates mutations in epigenetic pathways without lethal consequences.
- Many genetic tools exist to manipulate and observe epigenetic changes.
This makes Arabidopsis ideal for discovering mechanisms that might be too subtle or too harmful to study directly in mammals.
Broader Implications of the Study
Because the study demonstrates that DNA sequences can influence methylation patterns, researchers may eventually develop precise epigenetic editing tools. Unlike genetic editing, epigenetic editing can silence or activate genes without permanently altering DNA sequences. This could allow safer, reversible modifications in agriculture or biotechnology. For example, engineered DNA motifs could encourage plants to silence harmful elements or activate beneficial stress-response genes during drought or high temperatures.
In human health, although the exact mechanisms differ, the principle that DNA may encode signals guiding epigenetic marks could reshape how we think about diseases associated with methylation errors. If parallels exist in mammals, scientists may discover new ways to prevent or repair aberrant methylation linked to cancer or other developmental disorders.
The Salk Institute study represents a major advance because it answers a question that has lingered for decades: How do new epigenetic patterns begin? The answer is now much clearer—plants use specific DNA sequences, recognized by specialized transcription factors, to guide the placement of epigenetic marks. This not only fills a gap in our understanding but also opens the door to innovative technologies that could reshape biology, agriculture, and medicine.
Research Paper:
https://www.nature.com/articles/s41556-025-01808-5