New BASIC Technique Reveals How Genetic Variants Drive Neurodegenerative Disease Risk
Understanding how genetics shapes disease risk has been one of the biggest challenges in modern biomedical research, especially when it comes to complex brain disorders. While scientists have made enormous progress in identifying genetic variants linked to diseases, connecting those variants to real changes in gene activity inside specific brain cells has remained difficult. A new study published in Nature Communications introduces a powerful method that takes a major step toward solving this problem, particularly for neurodegenerative diseases such as Alzheimer’s disease and amyotrophic lateral sclerosis (ALS).
Researchers from Penn State College of Medicine have developed a technique that significantly improves how scientists map genetic variants to disease-related gene expression in the brain. Instead of treating brain cell types as completely separate units, this approach models how genetic effects are shared and distinct across multiple cell types, unlocking insights that were previously hidden.
Why Traditional Genetic Studies Fall Short
Many diseases are influenced not just by whether a gene exists, but by how much that gene is expressed and in which cell types. Genome-wide association studies (GWAS) have been extremely useful for identifying regions of the genome associated with disease risk. However, most of the variants discovered by GWAS do not directly alter genes themselves. Instead, they affect gene regulation, making it hard to determine which genes are actually involved and how they contribute to disease.
Another major limitation is that GWAS often rely on bulk tissue samples. In brain research, this means mixing together neurons, glial cells, microglia, and other cell types into one averaged signal. Important genetic effects occurring in rare cell populations can easily get lost in this mix.
Single-cell sequencing has helped address this issue by allowing scientists to analyze gene expression in individual cell types. But single-cell data come with their own challenges. Sample sizes are often small, and rare cell types—like microglia, the brain’s immune cells—are especially difficult to study with sufficient statistical power.
A New Way to Look at Brain Genetics
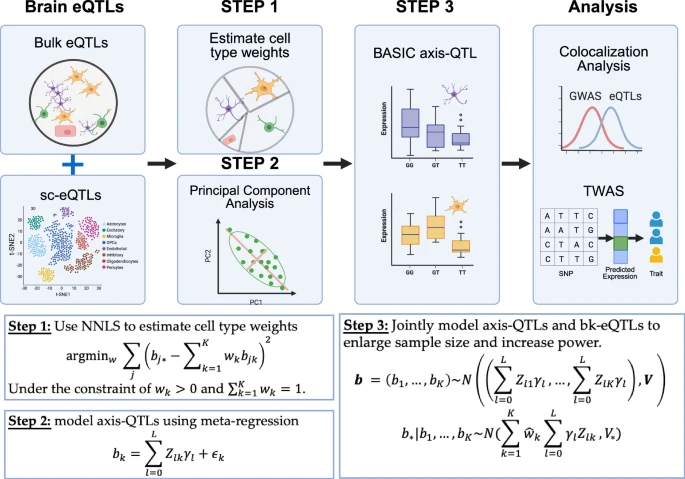
To overcome these limitations, the Penn State team developed a method called BASIC, which stands for Bulk And Single-cell eQTL Integration across Cell states. The key idea behind BASIC is simple but powerful: instead of analyzing each brain cell type independently, it models genetic effects that are shared across seven major brain cell types, while also capturing effects that are unique to specific cells.
This approach reflects biological reality more closely. Many genes that are essential for survival are active in more than one cell type. By identifying these shared regulatory patterns, BASIC can extract far more information from existing datasets without needing to collect dramatically more samples.
The researchers combined bulk tissue data, which provide large sample sizes, with single-cell data, which provide cell-type specificity. By integrating both sources, BASIC achieves the strengths of each while minimizing their weaknesses.
Dramatic Improvements in Gene Discovery
When the team compared BASIC to existing methods, the results were striking. The new approach identified approximately 75% more genes associated with genetic regulation than traditional single-cell methods. This improvement is equivalent to increasing the effective sample size by nearly 77%, without collecting additional data.
The researchers applied BASIC to genetic data from 12 brain-related conditions, including Alzheimer’s disease, ALS, schizophrenia, addiction, and other neurological and psychiatric disorders. Compared to single-cell analysis alone, BASIC improved the accuracy of linking genetic variants to disease-associated genes by more than 53%. When compared to bulk tissue analysis alone, the improvement exceeded 111%.
These gains are not just statistical. BASIC uncovered new genes linked to neurodegenerative disease risk that had been missed by conventional approaches, providing fresh leads for understanding disease mechanisms.
Insights Into Alzheimer’s Disease and ALS
Alzheimer’s disease has long been associated with genes such as APOE, which can increase disease risk by several-fold depending on the variant. However, APOE is only part of a much larger genetic network. The BASIC method helped identify additional genes whose expression patterns across multiple brain cell types are associated with Alzheimer’s risk.
Similarly, in ALS, a devastating neurodegenerative disease affecting motor neurons, BASIC revealed regulatory genes that had not previously been connected to disease risk. These discoveries help clarify how genetic variation influences disease development at the cellular level, rather than just pointing to broad genomic regions.
A particularly important aspect of this work is its ability to shed light on microglia, which play a key role in neuroinflammation. Neuroinflammation is increasingly recognized as a major contributor to neurodegenerative diseases, and understanding how genetic variants affect microglial function could open new therapeutic directions.
Implications for Drug Discovery and Repurposing
Beyond identifying disease-associated genes, the researchers explored how their findings could be used to guide treatment strategies. By examining gene expression patterns linked to disease risk, they searched for drug compounds capable of reversing those harmful expression profiles.
This analysis highlighted several FDA-approved drugs that could potentially be repurposed for brain disorders. For example, alfacalcidol, a synthetic form of vitamin D currently used for bone-related conditions, showed promise for reversing gene expression patterns associated with schizophrenia. Another drug, cabergoline, typically prescribed to treat high prolactin levels, emerged as a potential candidate for Alzheimer’s disease.
While these findings are preliminary and require extensive follow-up research, they demonstrate how improved genetic mapping can directly inform therapeutic discovery. Drug repurposing is especially attractive because approved medications already have known safety profiles, potentially speeding up the path to clinical application.
Why Shared Genetic Effects Matter
One of the most interesting ideas behind BASIC is its emphasis on shared genetic regulation across cell types. Instead of assuming that each cell type operates in isolation, the method acknowledges that biology often reuses the same regulatory mechanisms in different contexts.
Genes that are crucial for brain function tend to be expressed in multiple cell types, albeit at different levels. By modeling these shared effects, BASIC increases statistical power and improves biological interpretability. This shift in perspective could influence how future genetic studies are designed and analyzed.
A New Direction for Brain Disease Research
The researchers behind BASIC emphasize that the field has invested heavily in generating large datasets, but comparatively less effort has gone into developing smarter ways to analyze them. This new method shows that better analysis can unlock hidden value from data that already exist.
Rather than requiring massive new sequencing efforts, BASIC demonstrates how integrating and modeling data more intelligently can lead to major breakthroughs. This approach has the potential to reshape how scientists study not only neurodegenerative diseases, but also a wide range of complex traits influenced by genetics.
Looking Ahead
As genetic and single-cell datasets continue to grow, methods like BASIC will become increasingly important. By bridging the gap between genetic variants and their functional effects in specific brain cells, this technique brings researchers closer to understanding the true biological roots of neurodegenerative disease.
While more validation and experimental follow-up are needed, the study offers a compelling example of how computational innovation can drive progress in medicine. For patients and researchers alike, this represents a hopeful step toward more precise diagnoses, better treatments, and ultimately improved outcomes for devastating brain disorders.
Research paper: https://www.nature.com/articles/s41467-025-65643-w