Using Genetic Signals and AI Together Could Transform How Pneumonia Is Diagnosed and Reduce Antibiotic Overuse
Diagnosing pneumonia has always been one of the tougher challenges in modern medicine, especially in critically ill patients. Lung infections remain among the leading causes of death worldwide, yet doctors often have to make treatment decisions with incomplete or ambiguous information. A new study from researchers at the University of California, San Francisco (UCSF) suggests that combining genetic biomarkers with artificial intelligence could significantly improve diagnostic accuracy and dramatically reduce unnecessary antibiotic use.
The findings, published in Nature Communications in December 2025, show that pairing a host gene expression marker with a large language model analyzing medical records can outperform both experienced physicians and traditional diagnostic methods.
Why Pneumonia Is So Hard to Diagnose
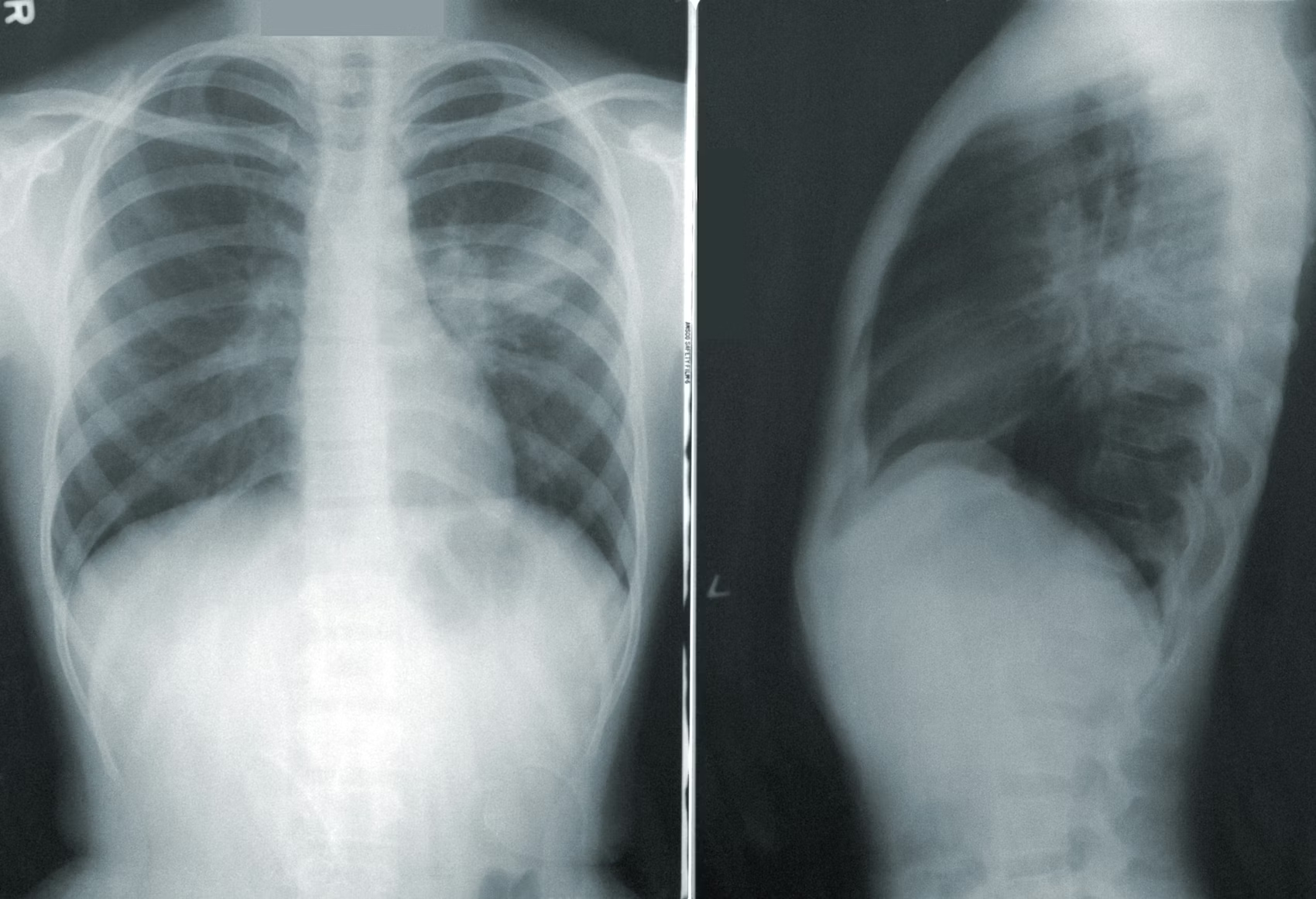
Pneumonia does not present the same way in every patient. Symptoms like shortness of breath, fever, low oxygen levels, and abnormal chest X-rays can also be caused by non-infectious conditions, such as fluid overload, inflammation, or lung injury. In intensive care units, where patients are already severely ill, this overlap makes diagnosis even harder.
Because of this uncertainty, clinicians often err on the side of caution and prescribe antibiotics “just in case.” While this approach can be lifesaving, it also contributes to antibiotic overuse, antibiotic resistance, and unnecessary side effects for patients who may not actually have an infection.
A New Diagnostic Strategy Combining Biology and AI
The UCSF research team explored a new way to address this problem by integrating two powerful tools:
- A host-response genetic biomarker known as FABP4
- A generative AI model (GPT-4) used to analyze electronic medical records
Instead of focusing solely on detecting pathogens like bacteria or viruses, the researchers looked at how the patient’s own immune system responds to infection. This approach is particularly useful when traditional cultures are slow or fail to identify a pathogen.
Understanding the FABP4 Biomarker
The biomarker at the center of this study is FABP4, a gene involved in regulating inflammation. Earlier work by the same team in 2023 showed that FABP4 behaves differently in infectious versus non-infectious lung conditions.
In simple terms, FABP4 expression is lower in immune cells during true lower respiratory tract infections compared to normal lung tissue. This difference creates a measurable signal that can help distinguish pneumonia from other causes of respiratory failure.
Because this biomarker reflects the host’s response, rather than the presence of a specific pathogen, it has the potential to work even when cultures are negative or delayed.
How AI Fits Into the Picture
The second component of the model involved GPT-4, deployed on a privacy-protecting, HIPAA-compliant platform developed at UCSF. The AI system analyzed unstructured clinical data, including:
- Physician notes
- Radiology reports
- Admission summaries
- Clinical histories
The AI was not trained to replace doctors, but rather to synthesize complex clinical information and assign a probability that a patient truly had a lower respiratory tract infection.
Interestingly, the researchers observed that the AI tended to place greater emphasis on radiology reports, while human physicians often focused more heavily on narrative clinical notes. This difference in weighting highlights how AI can provide a complementary perspective rather than simply mimicking human reasoning.
Study Design and Patient Groups
The study analyzed data from 157 critically ill adults, divided into two cohorts:
- 98 patients recruited before the COVID-19 pandemic, most of whom had bacterial infections
- 59 patients recruited during the pandemic, many of whom had viral infections, including COVID-19
This design allowed the researchers to test whether the model worked across different types of respiratory infections and clinical contexts.
Diagnostic Accuracy: Numbers That Stand Out
When tested individually, both components performed reasonably well:
- FABP4 biomarker alone: approximately 80% accuracy
- AI analysis alone: approximately 80% accuracy
However, when combined into a single diagnostic classifier, performance improved dramatically:
- Overall diagnostic accuracy: 96%
- Better distinction between infectious and non-infectious causes than ICU physicians at admission
The combined model consistently outperformed standard clinical diagnosis, particularly in identifying patients who did not actually have pneumonia.
Impact on Antibiotic Use
One of the most striking findings was the model’s potential effect on antibiotic prescribing. In the study population, clinicians prescribed antibiotics to most patients suspected of pneumonia.
Using retrospective analysis, the researchers estimated that if the biomarker-plus-AI model had been available at the time of admission, inappropriate antibiotic use could have been reduced by more than 80%.
This has major implications for:
- Antibiotic resistance, a growing global health crisis
- Reducing unnecessary drug exposure
- Improving antibiotic stewardship in hospitals
Comparing AI and Human Experts
To further validate the AI’s performance, the team compared GPT-4’s record analysis with evaluations from three physicians specializing in internal medicine and infectious diseases.
The results showed that:
- AI and physicians achieved similar overall accuracy
- AI relied more heavily on imaging data
- Physicians emphasized clinical narratives and judgment
Rather than viewing this as competition, the researchers emphasized that this difference demonstrates how AI can augment clinical decision-making by highlighting aspects of the data that humans may underweight.
Ease of Use and Transparency
Another notable aspect of the study is its accessibility. The research team published the AI prompts used in their analysis and emphasized that clinicians do not need advanced training in bioinformatics to use the approach.
The goal is to make the system simple to deploy, fast, and practical in real-world clinical settings.
Broader Context: Why Host-Response Diagnostics Matter
Traditional infectious disease diagnostics focus on identifying pathogens directly. While effective, these methods have limitations:
- Cultures can take days
- Pathogens are not always detectable
- Prior antibiotic use can obscure results
Host-response diagnostics, like FABP4, represent a shift toward understanding how the body reacts to infection, offering a faster and potentially more reliable signal.
When paired with AI capable of interpreting complex clinical data, this approach reflects a broader trend toward precision medicine, where diagnosis and treatment are tailored to individual patients rather than broad assumptions.
What Comes Next
The UCSF team is now working to validate the model as a clinical diagnostic test, moving beyond observational research. They also plan to apply a similar strategy to sepsis, another life-threatening condition that is notoriously difficult to diagnose early and accurately.
If successful, this approach could mark a significant step toward smarter diagnostics that reduce uncertainty, improve patient outcomes, and curb unnecessary treatments.
Research Reference
Hoang Van Phan et al., Integrating a host biomarker with a large language model for diagnosis of lower respiratory tract infection, Nature Communications (2025).
https://www.nature.com/articles/s41467-025-66218-5