How Scientists Used AI to Stop Herpes Viruses From Entering Human Cells
Washington State University researchers have uncovered a remarkably precise way to stop certain viruses from entering human cells, and the discovery could reshape how future antiviral treatments are developed. By focusing on a single critical molecular interaction inside a viral protein, the team demonstrated that disrupting just one well-chosen connection can prevent a virus from fusing with a cell and starting an infection.
The research centers on herpesviruses, a large family of viruses responsible for many common and lifelong infections. What makes this study stand out is not only what the scientists found, but how they found it—by combining artificial intelligence, molecular simulations, and lab experiments to cut through a massive amount of biological complexity.
Blocking Viral Entry at the Molecular Level
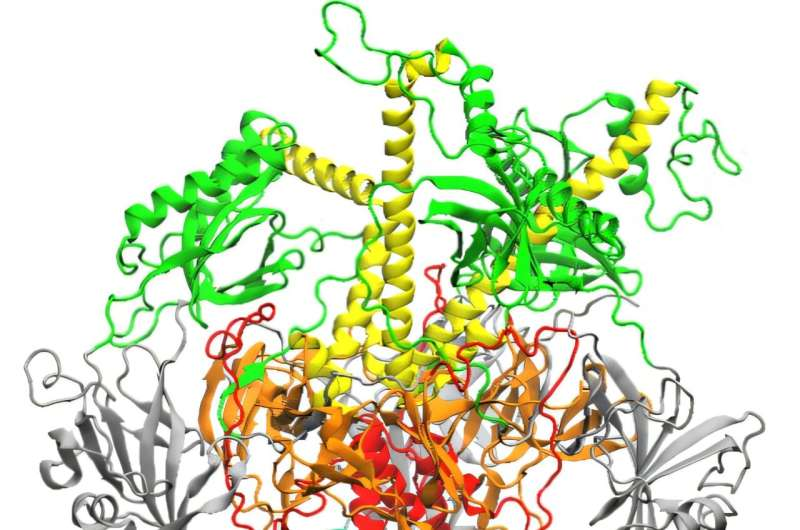
Viruses cannot reproduce on their own. To survive, they must first enter a host cell, hijack its machinery, and begin replication. For herpesviruses, that entry step depends on a sophisticated protein called glycoprotein B, often shortened to gB.
Glycoprotein B is known as a fusion protein. Its job is to help the virus merge its outer membrane with the membrane of a host cell. Once fusion happens, the virus gains access to the cell’s interior and infection begins. If fusion fails, the virus is essentially locked out.
The challenge is that gB is extremely complex. It is made up of hundreds of amino acids interacting with each other in countless ways. Scientists have long known that gB is essential for viral entry, but pinpointing exactly which interactions matter most has been difficult. Many interactions are present, but only a few are truly critical.
This is where the WSU team made a breakthrough.
Using AI to Find One Crucial Interaction
Researchers from WSU’s School of Mechanical and Materials Engineering and the Department of Veterinary Microbiology and Pathology used advanced molecular simulations and machine learning algorithms to analyze thousands of interactions inside glycoprotein B.
Instead of testing each interaction one by one in the lab—a process that could take years—the team built computational models that could rapidly evaluate which amino acid interactions were most important for fusion.
Their AI-based approach allowed them to sort through massive datasets and identify a single key interaction between two amino acids inside gB. This interaction helps stabilize the protein in a form that allows it to change shape and trigger membrane fusion.
Once that interaction was identified, the next step was to see what would happen if it were disrupted.
A Single Mutation With a Huge Effect
After the computational predictions were made, experimental work began under the leadership of researchers in veterinary microbiology and pathology. The team introduced a specific mutation into one of the amino acids involved in the critical interaction.
The result was striking.
With just that one change, the herpesvirus lost its ability to fuse with host cells. Without fusion, the virus could not enter the cell, and infection was effectively blocked. This confirmed that the AI-predicted interaction was not just important—it was essential.
What makes this especially impressive is that the researchers didn’t need to dismantle the entire protein or block multiple pathways. They simply modulated one interaction out of thousands, and that was enough to stop viral entry.
Why This Matters for Antiviral Research
Most existing antiviral drugs work after a virus has already entered cells, often by interfering with replication. While these treatments can be effective, they don’t stop the initial infection event.
This research points to a different strategy: preventing viral entry altogether. If a virus can’t get into a cell, it can’t replicate, spread, or cause disease.
Herpesviruses are particularly challenging because they can remain dormant in the body for years and reactivate later. A treatment that blocks entry could reduce the chance of initial infection or reinfection and may eventually complement vaccines or existing antivirals.
Even more importantly, glycoprotein B-like fusion proteins are found in many viruses, not just herpesviruses. That means the approach demonstrated here could inspire similar strategies against other viral pathogens.
Speeding Up Discovery With Computation
One of the biggest takeaways from this study is how powerful the combination of theory, simulation, and experiment can be.
Testing a single amino acid interaction experimentally can take months. Testing thousands through trial and error would be unrealistic. By narrowing the field using simulations and machine learning, the researchers dramatically accelerated the discovery process.
This approach represents a growing trend in biology and medicine, where AI helps scientists focus their experimental efforts on the most promising targets instead of searching blindly.
What Scientists Still Don’t Fully Understand
While the study clearly shows that disrupting one interaction can block fusion, there are still unanswered questions.
Researchers do not yet have a complete picture of how the mutation changes the overall structure of glycoprotein B at larger scales. Proteins are dynamic, constantly shifting their shapes, and understanding how a small local change influences the entire structure remains a challenge.
Future work will involve more advanced simulations and experiments to map how this specific interaction affects the protein’s behavior during the fusion process.
The Team Behind the Discovery
The research was led by faculty members from Washington State University, including experts in mechanical engineering, materials science, and veterinary microbiology. Graduate students played a key role in both the computational and experimental aspects of the project.
This interdisciplinary collaboration was essential. The problem required deep knowledge of physics, biology, computer science, and virology, all working together toward a single goal.
A Broader Look at Viral Fusion Proteins
Viral fusion proteins like glycoprotein B are among the most fascinating tools in nature. They act like molecular machines, sensing environmental cues and dramatically reshaping themselves to bring two membranes together.
Because fusion is such a delicate process, it often depends on a small number of highly specific interactions. This makes fusion proteins attractive targets for drugs—but also difficult to study.
The WSU study shows that with the right computational tools, it is possible to identify these weak points with surprising precision.
Looking Ahead
This research does not immediately translate into a new drug, but it lays critical groundwork. By proving that modulating a single molecular interaction can stop viral entry, the study opens the door to new antiviral strategies that are more targeted and potentially more effective.
As AI and molecular modeling tools continue to improve, similar approaches could help scientists uncover hidden vulnerabilities in other viruses and complex biological systems.
For now, this work stands as a powerful example of how modern computational science can uncover solutions to problems that once seemed overwhelmingly complex.
Research paper:
https://doi.org/10.1039/d5nr03235k