MARTi: The New Real-Time Software Revolutionizing Microbial Threat Detection
A team from the Earlham Institute in the UK has introduced an innovative software tool called MARTi—short for Metagenomic Analysis in Real-Time. This open-source platform is designed to rapidly identify and analyze microbial threats as DNA sequencing happens, significantly accelerating the process of detecting, monitoring, and responding to pathogens in clinical, environmental, agricultural, and biosecurity settings.
What Exactly Is MARTi?
MARTi is built to handle the demanding task of metagenomic analysis, a branch of genomics that studies the collection of genetic material from all organisms present in an environment—whether that’s soil, water, air, or the human body. Traditionally, analyzing metagenomic data can take hours or even days. MARTi changes that by enabling real-time analysis, meaning the system starts processing and identifying organisms as soon as sequencing begins.
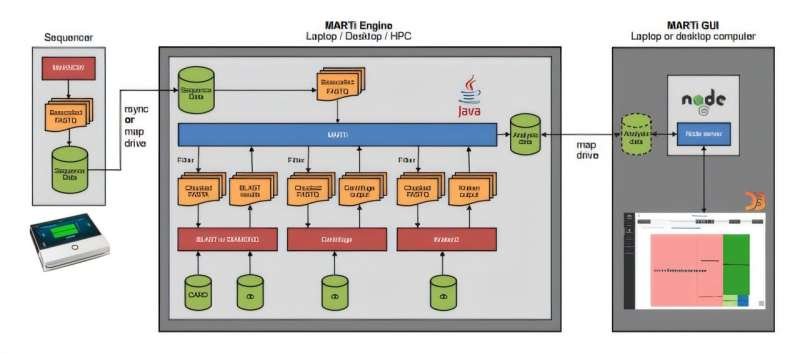
The tool is made up of two main components:
- MARTi Engine – A Java-based backend that handles the heavy lifting of data analysis. It processes DNA sequences, performs taxonomic classification (figuring out what species are present), calculates their abundance, and identifies antimicrobial resistance (AMR) genes or other key genetic features.
- MARTi GUI – A browser-based graphical interface that allows scientists to visualize and compare their results easily. Users can generate charts, graphs, and detailed visual summaries without writing a single line of code.
This setup gives MARTi remarkable flexibility. It can be deployed on a laptop for fieldwork—say, in a farm or research vessel—or scaled up to a high-performance computing (HPC) system for large and complex datasets.
Why Real-Time Metagenomics Matters
The concept of real-time metagenomics is simple but powerful: rather than waiting until all DNA sequencing is complete, analysis runs alongside sequencing. This makes it possible to detect threats as soon as they appear. For instance, if an unexpected pathogen is found in a hospital sample, clinicians could act within hours instead of days.
This kind of immediacy is game-changing in multiple domains:
- Clinical medicine: Hospitals could identify infections much faster, helping doctors choose targeted treatments sooner.
- Agriculture: Farmers could monitor for plant or livestock diseases before they spread.
- Environmental surveillance: Researchers could continuously sample water, soil, or air to track microbial shifts linked to pollution or ecosystem health.
- Biosecurity: Air and surface sampling in public spaces could quickly identify potential biothreats.
How MARTi Works in Practice
MARTi’s workflow integrates seamlessly with modern nanopore sequencing technologies, such as those from Oxford Nanopore Technologies (ONT). These devices are portable and capable of streaming sequence data in real time.
Once sequencing begins, the MARTi Engine takes over:
- It reads incoming data, filters out low-quality or short reads, and aligns the sequences against reference databases.
- It classifies the species present using tools like BLAST, Centrifuge, or Kraken2, depending on user preference.
- The results update dynamically, feeding into the GUI, where users can view interactive dashboards that display abundance charts, heatmaps, and treemaps showing microbial composition.
Crucially, MARTi supports custom databases, letting researchers tailor their analyses to specific projects—such as focusing on human pathogens, crop microbes, or marine species.
The system also includes built-in AMR gene detection, enabling the identification of antibiotic-resistant organisms directly from metagenomic data. Researchers can link these genes back to the species carrying them through a feature called taxon-AMR “walk-out”, which provides deeper insight into resistance pathways.
Built for the Field and the Lab
One of MARTi’s biggest advantages is how lightweight and portable it is. It doesn’t demand specialized hardware, and it can even run efficiently on a standard laptop—ideal for in-field use. This opens doors for practical applications like:
- Monitoring pathogens in remote regions (e.g., polar research expeditions or tropical rainforests).
- Running onboard sequencing during marine or environmental sampling missions.
- Supporting on-site diagnostics in low-resource settings.
The Earlham Institute has already demonstrated MARTi’s capabilities in projects such as AirSeq, a technology developed in partnership with the Natural History Museum, London. AirSeq captures and purifies DNA from air samples—then MARTi analyzes this data to detect airborne pathogens. The long-term vision is to have autonomous systems in agricultural fields, constantly sampling air and sending real-time alerts to farmers about emerging microbial threats.
Visuals and Usability
The MARTi GUI is not just an accessory—it’s central to how the system empowers users. Through its browser interface, users can:
- Access sample dashboards that display classification results and abundance plots.
- Compare multiple samples side-by-side to identify differences across environments or time points.
- Export publication-ready graphs and data summaries directly for reports or presentations.
Because it’s open-source and customizable, users can modify thresholds, visual preferences, or even integrate new analysis tools as needed. This flexibility makes MARTi suitable for both research laboratories and practical, field-level deployments.
Robust and Tested
The development team, led by Dr. Richard Leggett and Dr. Ned Peel, initially built MARTi for rapid pathogen identification in preterm infants—a clinical context where time is critical. Since then, the software has evolved into a general-purpose metagenomic platform used in a variety of research settings, from hospitals to agricultural and environmental studies.
Through extensive testing on both real and simulated datasets, MARTi has shown robust and reliable performance. It has now become the Earlham Institute’s default analysis tool for metagenomic projects.
Understanding Metagenomics and Its Importance
To appreciate MARTi’s impact, it’s worth revisiting what metagenomics actually involves. Traditional microbiology focuses on culturing individual organisms in the lab—a process that excludes most microbes because they’re difficult or impossible to grow. Metagenomics bypasses this limitation by sequencing all genetic material in a sample, allowing researchers to identify and study entire microbial communities at once.
Applications of metagenomics include:
- Health research: studying the human microbiome and its role in diseases like diabetes, obesity, and autoimmune disorders.
- Environmental monitoring: tracking microbial changes that signal pollution or climate change impacts.
- Agriculture: optimizing soil health by understanding beneficial and harmful microbes.
- Industrial biotechnology: discovering enzymes or biochemical pathways with commercial potential.
Real-time analysis tools like MARTi bring metagenomics closer to real-world decision-making—whether it’s stopping an outbreak or improving crop yields.
Future Prospects and Funding
The MARTi project received support from the UK Biotechnology and Biological Sciences Research Council (BBSRC) as part of the Norwich Research Park Biosciences Doctoral Training Partnership and other strategic programs. It’s part of a broader move toward accessible, portable bioinformatics solutions that can handle data at the point of collection.
Beyond research, MARTi’s architecture could be adapted for commercial or government biosurveillance systems. Some of its components may even find applications in defense and public health programs that need rapid threat detection.
As of 2025, the full research has been published in Genome Research, following its earlier release as a bioRxiv preprint. The developers have made it clear that the software will remain open-source, inviting collaboration and expansion by the wider scientific community.
Challenges and Limitations
Like any tool, MARTi isn’t without constraints. Its performance depends heavily on:
- The quality of input data—noisy or low-quality reads can reduce accuracy.
- The power of available hardware, especially for very large datasets.
- The completeness of reference databases, since uncharacterized organisms can’t be classified.
Despite these challenges, MARTi represents a major step toward democratizing metagenomic analysis. It combines scientific rigor with accessibility, allowing researchers—and potentially even field operators—to get actionable insights within hours.
Research Reference:
MARTi: a real-time analysis and visualisation tool for nanopore metagenomics (Genome Research, 2025)